The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells
- PMID: 15450134
- PMCID: PMC7091875
- DOI: 10.1038/sj.cr.7290240
The spike protein of severe acute respiratory syndrome (SARS) is cleaved in virus infected Vero-E6 cells
Abstract
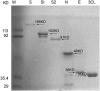
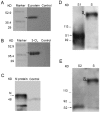
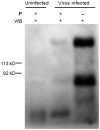
Spike protein is one of the major structural proteins of severe acute respiratory syndrome-coronavirus. It is essential for the interaction of the virons with host cell receptors and subsequent fusion of the viral envelop with host cell membrane to allow infection. Some spike proteins of coronavirus, such as MHV, HCoV-OC43, AIBV and BcoV, are proteolytically cleaved into two subunits, S1 and S2. In contrast, TGV, FIPV and HCoV-229E are not. Many studies have shown that the cleavage of spike protein seriously affects its function. In order to investigate the maturation and proteolytic processing of the S protein of SARS CoV, we generated S1 and S2 subunit specific antibodies (Abs) as well as N, E and 3CL protein-specific Abs. Our results showed that the antibodies could efficiently and specifically bind to their corresponding proteins from E.coli expressed or lysate of SARS-CoV infected Vero-E6 cells by Western blot analysis. Furthermore, the anti-S1 and S2 Abs were proved to be capable of binding to SARS CoV under electron microscope observation. When S2 Ab was used to perform immune precipitation with lysate of SARS-CoV infected cells, a cleaved S2 fragment was detected with S2-specific mAb by Western blot analysis. The data demonstrated that the cleavage of S protein was observed in the lysate, indicating that proteolytic processing of S protein is present in host cells.
Figures

Similar articles
-
Adenovirus-mediated expression of the C-terminal domain of SARS-CoV spike protein is sufficient to induce apoptosis in Vero E6 cells.FEBS Lett. 2005 Dec 19;579(30):6699-704. doi: 10.1016/j.febslet.2005.10.065. Epub 2005 Nov 21. FEBS Lett. 2005. PMID: 16310778 Free PMC article.
-
SARS-CoV spike proteins expressed by the vaccinia virus Tiantan strain: secreted sq protein induces robust neutralization antibody in mice.Viral Immunol. 2009 Feb;22(1):57-66. doi: 10.1089/vim.2008.0064. Viral Immunol. 2009. PMID: 19210229
-
Epitope mapping and biological function analysis of antibodies produced by immunization of mice with an inactivated Chinese isolate of severe acute respiratory syndrome-associated coronavirus (SARS-CoV).Virology. 2005 Mar 30;334(1):134-43. doi: 10.1016/j.virol.2005.01.035. Virology. 2005. PMID: 15749129 Free PMC article.
-
[Protease-dependent cell entry mechanism of coronaviruses].Uirusu. 2011 Jun;61(1):109-16. doi: 10.2222/jsv.61.109. Uirusu. 2011. PMID: 21972562 Review. Japanese.
-
[Cell entry mechanism of coronaviruses: implication in their pathogenesis].Uirusu. 2006 Dec;56(2):165-71. doi: 10.2222/jsv.56.165. Uirusu. 2006. PMID: 17446665 Review. Japanese.
Cited by
-
Screening and identification of B cell epitope of the nucleocapsid protein in SARS-CoV-2 using the monoclonal antibodies.Appl Microbiol Biotechnol. 2022 Feb;106(3):1151-1164. doi: 10.1007/s00253-022-11769-6. Epub 2022 Jan 17. Appl Microbiol Biotechnol. 2022. PMID: 35037999 Free PMC article.
-
Serological and molecular findings during SARS-CoV-2 infection: the first case study in Finland, January to February 2020.Euro Surveill. 2020 Mar;25(11):2000266. doi: 10.2807/1560-7917.ES.2020.25.11.2000266. Euro Surveill. 2020. PMID: 32209163 Free PMC article.
-
Synthesis and Identification of Novel Potential Molecules Against COVID-19 Main Protease Through Structure-Guided Virtual Screening Approach.Appl Biochem Biotechnol. 2021 Nov;193(11):3602-3623. doi: 10.1007/s12010-021-03615-8. Epub 2021 Jul 29. Appl Biochem Biotechnol. 2021. PMID: 34324152 Free PMC article.
-
Characterization and application of monoclonal antibodies against N protein of SARS-coronavirus.Biochem Biophys Res Commun. 2005 Oct 14;336(1):110-7. doi: 10.1016/j.bbrc.2005.08.032. Biochem Biophys Res Commun. 2005. PMID: 16112641 Free PMC article.
-
Molecular docking, DFT analysis, and dynamics simulation of natural bioactive compounds targeting ACE2 and TMPRSS2 dual binding sites of spike protein of SARS CoV-2.J Mol Liq. 2021 Nov 15;342:116942. doi: 10.1016/j.molliq.2021.116942. Epub 2021 Jul 9. J Mol Liq. 2021. PMID: 34305216 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

