An experimentally determined protein folding energy landscape
- PMID: 15377792
- PMCID: PMC521126
- DOI: 10.1073/pnas.0403386101
An experimentally determined protein folding energy landscape
Abstract
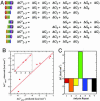
Energy landscapes have been used to conceptually describe and model protein folding but have been difficult to measure experimentally, in large part because of the myriad of partly folded protein conformations that cannot be isolated and thermodynamically characterized. Here we experimentally determine a detailed energy landscape for protein folding. We generated a series of overlapping constructs containing subsets of the seven ankyrin repeats of the Drosophila Notch receptor, a protein domain whose linear arrangement of modular structural units can be fragmented without disrupting structure. To a good approximation, stabilities of each construct can be described as a sum of energy terms associated with each repeat. The magnitude of each energy term indicates that each repeat is intrinsically unstable but is strongly stabilized by interactions with its nearest neighbors. These linear energy terms define an equilibrium free energy landscape, which shows an early free energy barrier and suggests preferred low-energy routes for folding.
Figures

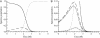
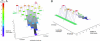
Similar articles
-
Studies of the ankyrin repeats of the Drosophila melanogaster Notch receptor. 2. Solution stability and cooperativity of unfolding.Biochemistry. 2001 Dec 4;40(48):14357-67. doi: 10.1021/bi011436+. Biochemistry. 2001. PMID: 11724547
-
Experimental characterization of the folding kinetics of the notch ankyrin domain.J Mol Biol. 2005 Sep 16;352(2):266-81. doi: 10.1016/j.jmb.2005.07.026. J Mol Biol. 2005. PMID: 16095609
-
Limits of cooperativity in a structurally modular protein: response of the Notch ankyrin domain to analogous alanine substitutions in each repeat.J Mol Biol. 2002 Nov 22;324(2):373-86. doi: 10.1016/s0022-2836(02)00945-2. J Mol Biol. 2002. PMID: 12441114
-
Folding landscapes of ankyrin repeat proteins: experiments meet theory.Curr Opin Struct Biol. 2008 Feb;18(1):27-34. doi: 10.1016/j.sbi.2007.12.004. Curr Opin Struct Biol. 2008. PMID: 18243686 Free PMC article. Review.
-
Evolution, energy landscapes and the paradoxes of protein folding.Biochimie. 2015 Dec;119:218-30. doi: 10.1016/j.biochi.2014.12.007. Epub 2014 Dec 18. Biochimie. 2015. PMID: 25530262 Free PMC article. Review.
Cited by
-
Highly polarized C-terminal transition state of the leucine-rich repeat domain of PP32 is governed by local stability.Proc Natl Acad Sci U S A. 2015 May 5;112(18):E2298-306. doi: 10.1073/pnas.1412165112. Epub 2015 Apr 20. Proc Natl Acad Sci U S A. 2015. PMID: 25902505 Free PMC article.
-
Salt dependence of an alpha-helical peptide folding energy landscapes.Biochemistry. 2009 Nov 17;48(45):10818-26. doi: 10.1021/bi9014709. Biochemistry. 2009. PMID: 19845367 Free PMC article.
-
The Right-Handed Parallel β-Helix Topology of Erwinia chrysanthemi Pectin Methylesterase Is Intimately Associated with Both Sequential Folding and Resistance to High Pressure.Biomolecules. 2021 Jul 22;11(8):1083. doi: 10.3390/biom11081083. Biomolecules. 2021. PMID: 34439750 Free PMC article.
-
Modulation of the multistate folding of designed TPR proteins through intrinsic and extrinsic factors.Protein Sci. 2012 Mar;21(3):327-38. doi: 10.1002/pro.2018. Protein Sci. 2012. PMID: 22170589 Free PMC article.
-
StaRProtein, a web server for prediction of the stability of repeat proteins.PLoS One. 2015 Mar 25;10(3):e0119417. doi: 10.1371/journal.pone.0119417. eCollection 2015. PLoS One. 2015. PMID: 25807112 Free PMC article.
References
-
- Matthews, C. R. (1993) Annu. Rev. Biochem. 62, 653–683. - PubMed
-
- Socci, N. D., Onuchic, J. N. & Wolynes, P. G. (1998) Proteins 32, 136–158. - PubMed
-
- Onuchic, J. N., Nymeyer, H., Garcia, A. E., Chahine, J. & Socci, N. D. (2000) Adv. Protein. Chem. 53, 87–152. - PubMed
-
- Veitshans, T., Klimov, D. & Thirumalai, D. (1997) Folding Des. 2, 1–22. - PubMed
-
- Dill, K. A. & Chan, H. S. (1997) Nat. Struct. Biol. 4, 10–19. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

