Abstract shapes of RNA
- PMID: 15371549
- PMCID: PMC519098
- DOI: 10.1093/nar/gkh779
Abstract shapes of RNA
Abstract
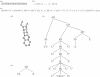
The function of a non-protein-coding RNA is often determined by its structure. Since experimental determination of RNA structure is time-consuming and expensive, its computational prediction is of great interest, and efficient solutions based on thermodynamic parameters are known. Frequently, however, the predicted minimum free energy structures are not the native ones, leading to the necessity of generating suboptimal solutions. While this can be accomplished by a number of programs, the user is often confronted with large outputs of similar structures, although he or she is interested in structures with more fundamental differences, or, in other words, with different abstract shapes. Here, we formalize the concept of abstract shapes and introduce their efficient computation. Each shape of an RNA molecule comprises a class of similar structures and has a representative structure of minimal free energy within the class. Shape analysis is implemented in the program RNAshapes. We applied RNAshapes to the prediction of optimal and suboptimal abstract shapes of several RNAs. For a given energy range, the number of shapes is considerably smaller than the number of structures, and in all cases, the native structures were among the top shape representatives. This demonstrates that the researcher can quickly focus on the structures of interest, without processing up to thousands of near-optimal solutions. We complement this study with a large-scale analysis of the growth behaviour of structure and shape spaces. RNAshapes is available for download and as an online version on the Bielefeld Bioinformatics Server.
Figures
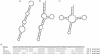

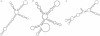

Similar articles
-
Structural analysis of aligned RNAs.Nucleic Acids Res. 2006;34(19):5471-81. doi: 10.1093/nar/gkl692. Epub 2006 Oct 4. Nucleic Acids Res. 2006. PMID: 17020924 Free PMC article.
-
pknotsRG: RNA pseudoknot folding including near-optimal structures and sliding windows.Nucleic Acids Res. 2007 Jul;35(Web Server issue):W320-4. doi: 10.1093/nar/gkm258. Epub 2007 May 3. Nucleic Acids Res. 2007. PMID: 17478505 Free PMC article.
-
Consensus shapes: an alternative to the Sankoff algorithm for RNA consensus structure prediction.Bioinformatics. 2005 Sep 1;21(17):3516-23. doi: 10.1093/bioinformatics/bti577. Epub 2005 Jul 14. Bioinformatics. 2005. PMID: 16020472
-
Sequence and structure analysis of noncoding RNAs.Methods Mol Biol. 2010;609:285-306. doi: 10.1007/978-1-60327-241-4_17. Methods Mol Biol. 2010. PMID: 20221926 Review.
-
Abstract shape analysis of RNA.Methods Mol Biol. 2014;1097:215-45. doi: 10.1007/978-1-62703-709-9_11. Methods Mol Biol. 2014. PMID: 24639162 Review.
Cited by
-
Thermodynamic matchers for the construction of the cuckoo RNA family.RNA Biol. 2015;12(2):197-207. doi: 10.1080/15476286.2015.1017206. RNA Biol. 2015. PMID: 25779873 Free PMC article.
-
Challenges and approaches to predicting RNA with multiple functional structures.RNA. 2018 Dec;24(12):1615-1624. doi: 10.1261/rna.067827.118. Epub 2018 Aug 24. RNA. 2018. PMID: 30143552 Free PMC article. Review.
-
PARTS: probabilistic alignment for RNA joinT secondary structure prediction.Nucleic Acids Res. 2008 Apr;36(7):2406-17. doi: 10.1093/nar/gkn043. Epub 2008 Feb 26. Nucleic Acids Res. 2008. PMID: 18304945 Free PMC article.
-
RNA structure prediction from evolutionary patterns of nucleotide composition.Nucleic Acids Res. 2009 Apr;37(5):1378-86. doi: 10.1093/nar/gkn987. Epub 2009 Jan 7. Nucleic Acids Res. 2009. PMID: 19129237 Free PMC article.
-
Phenotype Bias Determines How Natural RNA Structures Occupy the Morphospace of All Possible Shapes.Mol Biol Evol. 2022 Jan 7;39(1):msab280. doi: 10.1093/molbev/msab280. Mol Biol Evol. 2022. PMID: 34542628 Free PMC article.
References
-
- Mathews D.H., Sabina,J., Zuker,M. and Turner,D.H. (1999) Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol., 288, 911–940. - PubMed
-
- McCaskill J.S. (1990) The equilibrium partition function and base pair binding probabilities for RNA secondary structure. Biopolymers, 29, 1105–1119. - PubMed
-
- Zuker M. (1989) On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52. - PubMed
-
- Wuchty S., Fontana,W., Hofacker,I.L. and Schuster,P. (1999) Complete suboptimal folding of RNA and the stability of secondary structures. Biopolymers, 49, 145–169. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

