A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe
- PMID: 15371329
- PMCID: PMC522986
- DOI: 10.1101/gad.1218004
A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe
Abstract
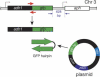
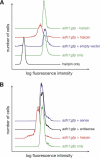
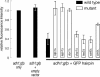
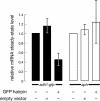
The Schizosaccharomyces pombe genome encodes only one of each of the three major classes of proteins implicated in RNA silencing: Dicer (Dcr1), RNA-dependent RNA polymerase (RdRP; Rdp1), and Argonaute (Ago1). These three proteins are required for silencing at centromeres and for the initiation of transcriptionally silent heterochromatin at the mating-type locus. Here, we show that the introduction of a double-stranded RNA (dsRNA) hairpin corresponding to a green fluorescent protein (GFP) transgene triggers classical RNA interference (RNAi) in S. pombe. That is, GFP silencing triggered by dsRNA reflects a change in the steady-state concentration of GFP mRNA, but not in the rate of GFP transcription. RNAi in S. pombe requires dcr1, rdp1, and ago1, but does not require chp1, tas3, or swi6, genes required for transcriptional silencing. Thus, the RNAi machinery in S. pombe can direct both transcriptional and posttranscriptional silencing using a single Dicer, RdRP, and Argonaute protein. Our findings suggest that these three proteins fulfill a common biochemical function in distinct siRNA-directed silencing pathways.
Copyright 2004 Cold Spring Harbor Laboratory Press
Figures
Similar articles
-
RNA interference (RNAi)-dependent and RNAi-independent association of the Chp1 chromodomain protein with distinct heterochromatic loci in fission yeast.Mol Cell Biol. 2005 Mar;25(6):2331-46. doi: 10.1128/MCB.25.6.2331-2346.2005. Mol Cell Biol. 2005. PMID: 15743828 Free PMC article.
-
The Chp1-Tas3 core is a multifunctional platform critical for gene silencing by RITS.Nat Struct Mol Biol. 2011 Nov 13;18(12):1351-7. doi: 10.1038/nsmb.2151. Nat Struct Mol Biol. 2011. PMID: 22081013 Free PMC article.
-
RNA interference effector proteins localize to mobile cytoplasmic puncta in Schizosaccharomyces pombe.Traffic. 2006 Aug;7(8):1032-44. doi: 10.1111/j.1600-0854.2006.00441.x. Epub 2006 May 25. Traffic. 2006. PMID: 16734665
-
Studies on the mechanism of RNAi-dependent heterochromatin assembly.Cold Spring Harb Symp Quant Biol. 2006;71:461-71. doi: 10.1101/sqb.2006.71.044. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381328 Review.
-
RNA interference and heterochromatin in the fission yeast Schizosaccharomyces pombe.Trends Genet. 2005 Aug;21(8):450-6. doi: 10.1016/j.tig.2005.06.005. Trends Genet. 2005. PMID: 15979194 Review.
Cited by
-
A Glimpse of "Dicer Biology" Through the Structural and Functional Perspective.Front Mol Biosci. 2021 May 7;8:643657. doi: 10.3389/fmolb.2021.643657. eCollection 2021. Front Mol Biosci. 2021. PMID: 34026825 Free PMC article. Review.
-
Exploring the functions of RNA interference pathway proteins: some functions are more RISCy than others?Biochem J. 2005 May 1;387(Pt 3):561-71. doi: 10.1042/BJ20041822. Biochem J. 2005. PMID: 15845026 Free PMC article. Review.
-
Heterochromatin assembly: a new twist on an old model.Chromosome Res. 2006;14(1):83-94. doi: 10.1007/s10577-005-1018-1. Chromosome Res. 2006. PMID: 16506098 Review.
-
On the origin and functions of RNA-mediated silencing: from protists to man.Curr Genet. 2006 Aug;50(2):81-99. doi: 10.1007/s00294-006-0078-x. Epub 2006 May 12. Curr Genet. 2006. PMID: 16691418 Free PMC article. Review.
-
Dicer associates with chromatin to repress genome activity in Schizosaccharomyces pombe.Nat Struct Mol Biol. 2011 Jan;18(1):94-9. doi: 10.1038/nsmb.1935. Epub 2010 Dec 12. Nat Struct Mol Biol. 2011. PMID: 21151114
References
-
- Aravin A.A., Naumova, N.M., Tulin, A.V., Vagin, V.V., Rozovsky, Y.M., and Gvozdev, V.A. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 11: 1017-1027. - PubMed
-
- Bahler J., Wu, J.Q., Longtine, M.S., Shah, N.G., McKenzie III, A., Steever, A.B., Wach, A., Philippsen, P., and Pringle, J.R. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943-951. - PubMed
-
- Bernstein E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363-366. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
