Vaccinia virus mutants with alanine substitutions in the conserved G5R gene fail to initiate morphogenesis at the nonpermissive temperature
- PMID: 15367589
- PMCID: PMC516429
- DOI: 10.1128/JVI.78.19.10238-10248.2004
Vaccinia virus mutants with alanine substitutions in the conserved G5R gene fail to initiate morphogenesis at the nonpermissive temperature
Abstract
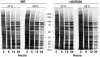
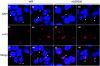
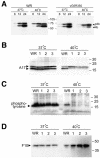
The initial characterization of the product of the vaccinia virus G5R gene, which is conserved in all poxviruses sequenced to date, is described. The G5 protein was detected in the core fraction of purified virions, and transcription and translation of the G5R open reading frame occurred early in infection, independently of DNA replication. Attempts to delete the G5R gene and isolate a replication-competent virus were unsuccessful, suggesting that G5R encodes an essential function. We engineered vaccinia virus mutants with clusters of charged amino acids changed to alanines and determined that several were unable to replicate at 40 degrees C but grew well at 37 degrees C. At the nonpermissive temperature, viral gene expression and DNA replication and processing were unperturbed. However, tyrosine phosphorylation and proteolytic cleavage of the A17 membrane protein and proteolytic cleavage of core proteins were inhibited at 40 degrees C, suggesting an assembly defect. The cytoplasm of cells that had been infected at the nonpermissive temperature contained large granular areas devoid of cellular organelles or virus structures except for occasional short crescent-shaped membranes and electron-dense lacy structures. The temperature-sensitive phenotype of the G5R mutants closely resembled the phenotypes of vaccinia virus mutants carrying conditionally lethal F10R protein kinase and H5R mutations. F10, although required for phosphorylation of A17 and viral membrane formation, was synthesized by the G5R mutants under nonpermissive conditions. An intriguing possibility is that G5 participates in the formation of viral membranes, a poorly understood event in poxvirus assembly.
Figures





Similar articles
-
Role of vaccinia virus A20R protein in DNA replication: construction and characterization of temperature-sensitive mutants.J Virol. 2001 Feb;75(4):1656-63. doi: 10.1128/JVI.75.4.1656-1663.2001. J Virol. 2001. PMID: 11160663 Free PMC article.
-
Vaccinia virus nonstructural protein encoded by the A11R gene is required for formation of the virion membrane.J Virol. 2005 Jun;79(11):6598-609. doi: 10.1128/JVI.79.11.6598-6609.2005. J Virol. 2005. PMID: 15890898 Free PMC article.
-
A complex of seven vaccinia virus proteins conserved in all chordopoxviruses is required for the association of membranes and viroplasm to form immature virions.Virology. 2004 Dec 20;330(2):447-59. doi: 10.1016/j.virol.2004.10.008. Virology. 2004. PMID: 15567438
-
Vaccinia virus DNA replication: a short review.Biochimie. 1995;77(10):774-9. doi: 10.1016/0300-9084(96)88195-8. Biochimie. 1995. PMID: 8824774 Review.
-
Poxvirus membrane biogenesis.Virology. 2015 May;479-480:619-26. doi: 10.1016/j.virol.2015.02.003. Epub 2015 Feb 26. Virology. 2015. PMID: 25728299 Free PMC article. Review.
Cited by
-
Vaccinia virus A6L encodes a virion core protein required for formation of mature virion.J Virol. 2007 Feb;81(3):1433-43. doi: 10.1128/JVI.02206-06. Epub 2006 Nov 15. J Virol. 2007. PMID: 17108027 Free PMC article.
-
Vaccinia virus A6 is essential for virion membrane biogenesis and localization of virion membrane proteins to sites of virion assembly.J Virol. 2012 May;86(10):5603-13. doi: 10.1128/JVI.00330-12. Epub 2012 Mar 7. J Virol. 2012. PMID: 22398288 Free PMC article.
-
Vaccinia virus L2 protein associates with the endoplasmic reticulum near the growing edge of crescent precursors of immature virions and stabilizes a subset of viral membrane proteins.J Virol. 2011 Dec;85(23):12431-41. doi: 10.1128/JVI.05573-11. Epub 2011 Sep 14. J Virol. 2011. PMID: 21917978 Free PMC article.
-
Poxvirus proteomics and virus-host protein interactions.Microbiol Mol Biol Rev. 2009 Dec;73(4):730-49. doi: 10.1128/MMBR.00026-09. Microbiol Mol Biol Rev. 2009. PMID: 19946139 Free PMC article. Review.
-
Participation of vaccinia virus l2 protein in the formation of crescent membranes and immature virions.J Virol. 2011 Mar;85(6):2504-11. doi: 10.1128/JVI.02505-10. Epub 2011 Jan 12. J Virol. 2011. PMID: 21228235 Free PMC article.
References
-
- Banham, A., and G. L. Smith. 1992. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology 191:803-812. - PubMed
-
- Baroudy, B. M., S. Venkatesan, and B. Moss. 1982. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp. Quant. Biol. 47:723-729. - PubMed
-
- Black, E. P., N. Moussatche, and R. C. Condit. 1998. Characterization of the interactions among vaccinia virus transcription factors G2R, A18R, and H5R. Virology 245:313-322. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

