Heterogeneous nuclear ribonucleoprotein K represses transcription from a cytosine/thymidine-rich element in the osteocalcin promoter
- PMID: 15361071
- PMCID: PMC1134736
- DOI: 10.1042/BJ20040680
Heterogeneous nuclear ribonucleoprotein K represses transcription from a cytosine/thymidine-rich element in the osteocalcin promoter
Abstract
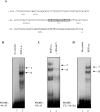
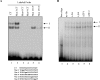
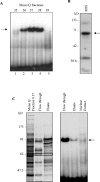
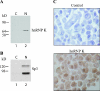
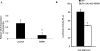
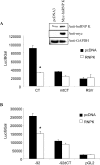
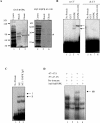
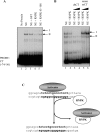
HnRNP K (heterogeneous nuclear ribonucleoprotein K) was biochemically purified from a screen of proteins co-purifying with binding activity to the osteocalcin promoter. We identify hnRNP K as a novel repressor of osteocalcin gene transcription. Overexpression of hnRNP K lowers the expression of osteocalcin mRNA by 5-fold. Furthermore, luciferase reporter assays demonstrate that overexpression of hnRNP K represses osteocalcin transcription from a CT (cytosine/thymidine)-rich element in the proximal promoter. Electrophoretic mobility-shift analysis reveals that recombinant hnRNP K binds to the CT-rich element, but binds ss (single-stranded), rather than ds (double-stranded) oligonucleotide probes. Accordingly, hnRNP K antibody can supershift a binding activity present in nuclear extracts using ss sense, but not antisense or ds oligonucleotides corresponding to the CT-rich -95 to -47 osteocalcin promoter. Importantly, addition of recombinant hnRNP K to ROS 17/2.8 nuclear extract disrupts formation of a DNA-protein complex on ds CT element oligonucleotides. This action is mutually exclusive with hnRNP K's ability to bind ss DNA. These results demonstrate that hnRNPK, although co-purified with a dsDNA-binding activity, does not itself bind dsDNA. Rather, hnRNP K represses osteocalcin gene transcription by inhibiting the formation of a transcriptional complex on the CT element of the osteocalcin promoter.
Figures
Similar articles
-
Transcriptional regulation of heterogeneous nuclear ribonucleoprotein K gene expression.Biochimie. 2015 Feb;109:27-35. doi: 10.1016/j.biochi.2014.12.002. Epub 2014 Dec 10. Biochimie. 2015. PMID: 25497182 Free PMC article.
-
Heterogeneous nuclear ribonucleoprotein K is a DNA-binding transactivator.J Biol Chem. 1995 Mar 3;270(9):4875-81. doi: 10.1074/jbc.270.9.4875. J Biol Chem. 1995. PMID: 7876260
-
Heterogeneous nuclear ribonucleoprotein K is a transcription factor.Mol Cell Biol. 1996 May;16(5):2350-60. doi: 10.1128/MCB.16.5.2350. Mol Cell Biol. 1996. PMID: 8628302 Free PMC article.
-
Identification of the SRC pyrimidine-binding protein (SPy) as hnRNP K: implications in the regulation of SRC1A transcription.Nucleic Acids Res. 2003 Mar 1;31(5):1502-13. doi: 10.1093/nar/gkg246. Nucleic Acids Res. 2003. PMID: 12595559 Free PMC article.
-
Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression.Cancer Lett. 2014 Oct 1;352(2):152-9. doi: 10.1016/j.canlet.2014.06.019. Epub 2014 Jul 10. Cancer Lett. 2014. PMID: 25016060 Review.
Cited by
-
Long noncoding RNA EWSAT1-mediated gene repression facilitates Ewing sarcoma oncogenesis.J Clin Invest. 2014 Dec;124(12):5275-90. doi: 10.1172/JCI72124. Epub 2014 Nov 17. J Clin Invest. 2014. PMID: 25401475 Free PMC article.
-
Interaction of connexin43 and protein kinase C-delta during FGF2 signaling.BMC Biochem. 2010 Mar 25;11:14. doi: 10.1186/1471-2091-11-14. BMC Biochem. 2010. PMID: 20338032 Free PMC article.
-
RNA-binding proteins and translational regulation in axons and growth cones.Front Neurosci. 2013 May 23;7:81. doi: 10.3389/fnins.2013.00081. eCollection 2013. Front Neurosci. 2013. PMID: 23734093 Free PMC article.
-
hnRNPK-derived cell-penetratingpeptide inhibits cancer cell survival.Mol Ther Oncolytics. 2021 Oct 19;23:342-354. doi: 10.1016/j.omto.2021.10.004. eCollection 2021 Dec 17. Mol Ther Oncolytics. 2021. PMID: 34820504 Free PMC article.
-
Genomic localization of RNA binding proteins reveals links between pre-mRNA processing and transcription.Genome Res. 2006 Jul;16(7):912-21. doi: 10.1101/gr.5211806. Epub 2006 Jun 12. Genome Res. 2006. PMID: 16769980 Free PMC article.
References
-
- Romberg R. W., Werness P. G., Riggs B. L., Mann K. G. Inhibition of hydroxyapatite crystal growth by bone-specific and other calcium-binding proteins. Biochemistry. 1986;25:1176–1180. - PubMed
-
- Hoang Q. Q., Sicheri F., Howard A. J., Yang D. S. Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature (London) 2003;425:977–980. - PubMed
-
- Ducy P., Zhang R., Geoffroy V., Ridall A. L., Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell (Cambridge, Mass.) 1997;89:747–754. - PubMed
-
- Towler D. A., Rutledge S.-J. C., Rodan G. A. Msx-2/Hox 8.1: a transcriptional regulator of the rat osteocalcin promoter. Mol. Endocrinol. 1994;8:1484–1493. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

