Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins
- PMID: 15359280
- PMCID: PMC522791
- DOI: 10.1038/sj.emboj.7600389
Essential role of Mia40 in import and assembly of mitochondrial intermembrane space proteins
Abstract
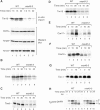
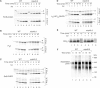
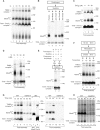
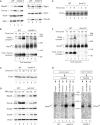
Mitochondria import nuclear-encoded precursor proteins to four different subcompartments. Specific import machineries have been identified that direct the precursor proteins to the mitochondrial outer membrane, inner membrane or matrix, respectively. However, a machinery dedicated to the import of mitochondrial intermembrane space (IMS) proteins has not been found so far. We have identified the essential IMS protein Mia40 (encoded by the Saccharomyces cerevisiae open reading frame YKL195w). Mitochondria with a mutant form of Mia40 are selectively inhibited in the import of several small IMS proteins, including the essential proteins Tim9 and Tim10. The import of proteins to the other mitochondrial subcompartments does not depend on functional Mia40. The binding of small Tim proteins to Mia40 is crucial for their transport across the outer membrane and represents an initial step in their assembly into IMS complexes. We conclude that Mia40 is a central component of the protein import and assembly machinery of the mitochondrial IMS.
Figures


Similar articles
-
Oxidative folding of small Tims is mediated by site-specific docking onto Mia40 in the mitochondrial intermembrane space.Mol Microbiol. 2007 Sep;65(5):1360-73. doi: 10.1111/j.1365-2958.2007.05880.x. Epub 2007 Aug 6. Mol Microbiol. 2007. PMID: 17680986
-
The essential mitochondrial protein Erv1 cooperates with Mia40 in biogenesis of intermembrane space proteins.J Mol Biol. 2005 Oct 28;353(3):485-92. doi: 10.1016/j.jmb.2005.08.051. Epub 2005 Sep 8. J Mol Biol. 2005. PMID: 16181637
-
Mia40, a novel factor for protein import into the intermembrane space of mitochondria is able to bind metal ions.FEBS Lett. 2005 Jan 3;579(1):179-84. doi: 10.1016/j.febslet.2004.11.072. FEBS Lett. 2005. PMID: 15620710
-
Diverse mechanisms and machineries for import of mitochondrial proteins.Biol Chem. 2007 Sep;388(9):891-7. doi: 10.1515/BC.2007.097. Biol Chem. 2007. PMID: 17696772 Review.
-
Protein import into mitochondria.IUBMB Life. 2001 Sep-Nov;52(3-5):101-12. doi: 10.1080/15216540152845894. IUBMB Life. 2001. PMID: 11798021 Review.
Cited by
-
Function and redox state of mitochondrial localized cysteine-rich proteins important in the assembly of cytochrome c oxidase.Biochim Biophys Acta. 2008 Apr;1783(4):618-28. doi: 10.1016/j.bbamcr.2007.10.016. Epub 2007 Nov 9. Biochim Biophys Acta. 2008. PMID: 18070608 Free PMC article. Review.
-
Structure of the mitochondrial import gate reveals distinct preprotein paths.Nature. 2019 Nov;575(7782):395-401. doi: 10.1038/s41586-019-1680-7. Epub 2019 Oct 10. Nature. 2019. PMID: 31600774
-
ALR and liver regeneration.Mol Cell Biochem. 2006 Aug;288(1-2):159-69. doi: 10.1007/s11010-006-9133-7. Epub 2006 May 12. Mol Cell Biochem. 2006. PMID: 16691313 Review.
-
DMR1 (CCM1/YGR150C) of Saccharomyces cerevisiae encodes an RNA-binding protein from the pentatricopeptide repeat family required for the maintenance of the mitochondrial 15S ribosomal RNA.Genetics. 2010 Apr;184(4):959-73. doi: 10.1534/genetics.110.113969. Epub 2010 Feb 1. Genetics. 2010. PMID: 20124025 Free PMC article.
-
Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins.Mol Biol Cell. 2006 Mar;17(3):1436-50. doi: 10.1091/mbc.e05-08-0740. Epub 2006 Jan 11. Mol Biol Cell. 2006. PMID: 16407407 Free PMC article.
References
-
- Allen S, Lu H, Thornton D, Tokatlidis K (2003) Juxtaposition of the two distal Cx3C motifs via intrachain disulfide bonding is essential for the folding of Tim10. J Biol Chem 278: 38505–38513 - PubMed
-
- Beers J, Glerum DM, Tzagoloff A (1997) Purification, characterization, and localization of yeast Cox17p, a mitochondrial copper shuttle. J Biol Chem 272: 33191–33196 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

