Over 20% of human transcripts might form sense-antisense pairs
- PMID: 15356298
- PMCID: PMC519112
- DOI: 10.1093/nar/gkh818
Over 20% of human transcripts might form sense-antisense pairs
Abstract
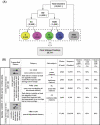
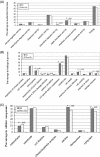
The major challenge to identifying natural sense- antisense (SA) transcripts from public databases is how to determine the correct orientation for an expressed sequence, especially an expressed sequence tag sequence. In this study, we established a set of very stringent criteria to identify the correct orientation of each human transcript. We used these orientation-reliable transcripts to create 26 741 transcription clusters in the human genome. Our analysis shows that 22% (5880) of the human transcription clusters form SA pairs, higher than any previous estimates. Our orientation-specific RT-PCR results along with the comparison of experimental data from previous studies confirm that our SA data set is reliable. This study not only demonstrates that our criteria for the prediction of SA transcripts are efficient, but also provides additional convincing data to support the view that antisense transcription is quite pervasive in the human genome. In-depth analyses show that SA transcripts have some significant differences compared with other types of transcripts, with regard to chromosomal distribution and Gene Ontology-annotated categories of physiological roles, functions and spatial localizations of gene products.
Figures

Similar articles
-
Detection of antisense RNA transcripts by strand-specific RT-PCR.Methods Mol Biol. 2010;630:125-38. doi: 10.1007/978-1-60761-629-0_9. Methods Mol Biol. 2010. PMID: 20300995
-
Sense and antisense transcripts of convergent gene pairs in Arabidopsis thaliana can share a common polyadenylation region.PLoS One. 2011 Feb 2;6(2):e16769. doi: 10.1371/journal.pone.0016769. PLoS One. 2011. PMID: 21311762 Free PMC article.
-
A genome-wide investigation of expression characteristics of natural antisense transcripts in liver and muscle samples of pigs.PLoS One. 2012;7(12):e52433. doi: 10.1371/journal.pone.0052433. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23285040 Free PMC article.
-
Making sense of the natural antisense transcript puzzle.Trends Plant Sci. 2021 Nov;26(11):1104-1115. doi: 10.1016/j.tplants.2021.07.004. Epub 2021 Jul 21. Trends Plant Sci. 2021. PMID: 34303604 Review.
-
Systematic search for natural antisense transcripts in eukaryotes (review).Int J Mol Med. 2005 Feb;15(2):197-203. Int J Mol Med. 2005. PMID: 15647831 Review.
Cited by
-
Non-coding rRNA-mediated preferential killing in cancer cells is enhanced by suppression of autophagy in non-transformed counterpart.Cell Death Dis. 2011 Dec 8;2(12):e239. doi: 10.1038/cddis.2011.110. Cell Death Dis. 2011. PMID: 22158478 Free PMC article.
-
Natural antisense transcript of natriuretic peptide precursor A (NPPA): structural organization and modulation of NPPA expression.BMC Mol Biol. 2009 Aug 11;10:81. doi: 10.1186/1471-2199-10-81. BMC Mol Biol. 2009. PMID: 19671135 Free PMC article.
-
Uncovering information on expression of natural antisense transcripts in Affymetrix MOE430 datasets.BMC Genomics. 2007 Jun 28;8:200. doi: 10.1186/1471-2164-8-200. BMC Genomics. 2007. PMID: 17598913 Free PMC article.
-
Systematic analysis of head-to-head gene organization: evolutionary conservation and potential biological relevance.PLoS Comput Biol. 2006 Jul 7;2(7):e74. doi: 10.1371/journal.pcbi.0020074. Epub 2006 May 15. PLoS Comput Biol. 2006. PMID: 16839196 Free PMC article.
-
Evidence for a major role of antisense RNAs in cyanobacterial gene regulation.Mol Syst Biol. 2009;5:305. doi: 10.1038/msb.2009.63. Epub 2009 Sep 15. Mol Syst Biol. 2009. PMID: 19756044 Free PMC article.
References
-
- Simons R.W. (1988) Naturally occurring antisense RNA control—a brief review. Gene, 72, 35–44. - PubMed
-
- Wagner E.G. and Simons,R.W. (1994) Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol., 48, 713–742. - PubMed
-
- Lacatena R.M. and Cesareni,G. (1981) Base pairing of RNA I with its complementary sequence in the primer precursor inhibits ColE1 replication. Nature, 294, 623–626. - PubMed
-
- Knee R. and Murphy,P.R. (1997) Regulation of gene expression by natural antisense RNA transcripts. Neurochem. Int., 31, 379–392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

