Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core
- PMID: 15345712
- PMCID: PMC8008698
- DOI: 10.1074/jbc.M408782200
Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core
Abstract
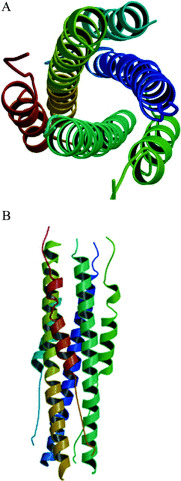
Severe acute respiratory syndrome coronavirus is a newly emergent virus responsible for a recent outbreak of an atypical pneumonia. The coronavirus spike protein, an enveloped glycoprotein essential for viral entry, belongs to the class I fusion proteins and is characterized by the presence of two heptad repeat (HR) regions, HR1 and HR2. These two regions are understood to form a fusion-active conformation similar to those of other typical viral fusion proteins. This hairpin structure likely juxtaposes the viral and cellular membranes, thus facilitating membrane fusion and subsequent viral entry. The fusion core protein of severe acute respiratory syndrome coronavirus spike protein was crystallized, and the structure was determined at 2.8 A of resolution. The fusion core is a six-helix bundle with three HR2 helices packed against the hydrophobic grooves on the surface of central coiled coil formed by three parallel HR1 helices in an oblique antiparallel manner. This structure shares significant similarity with the fusion core structure of mouse hepatitis virus spike protein and other viral fusion proteins, suggesting a conserved mechanism of membrane fusion. Drug discovery strategies aimed at inhibiting viral entry by blocking hairpin formation, which have been successfully used in human immunodeficiency virus 1 inhibitor development, may be applicable to the inhibition of severe acute respiratory syndrome coronavirus on the basis of structural information provided here. The relatively deep grooves on the surface of the central coiled coil will be a good target site for the design of viral fusion inhibitors.
Figures



Similar articles
-
Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core.J Biol Chem. 2004 Jul 16;279(29):30514-22. doi: 10.1074/jbc.M403760200. Epub 2004 Apr 27. J Biol Chem. 2004. PMID: 15123674 Free PMC article.
-
Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus.Biochemistry. 2004 Nov 9;43(44):14064-71. doi: 10.1021/bi049101q. Biochemistry. 2004. PMID: 15518555
-
Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein.Proc Natl Acad Sci U S A. 2004 Dec 28;101(52):17958-63. doi: 10.1073/pnas.0406128102. Epub 2004 Dec 16. Proc Natl Acad Sci U S A. 2004. PMID: 15604146 Free PMC article.
-
Severe acute respiratory syndrome coronavirus entry into host cells: Opportunities for therapeutic intervention.Med Res Rev. 2006 Jul;26(4):414-33. doi: 10.1002/med.20055. Med Res Rev. 2006. PMID: 16521129 Free PMC article. Review.
-
The SARS-CoV S glycoprotein.Cell Mol Life Sci. 2004 Oct;61(19-20):2428-30. doi: 10.1007/s00018-004-4257-y. Cell Mol Life Sci. 2004. PMID: 15526150 Free PMC article. Review.
Cited by
-
Sulfated Glycans Inhibit the Interaction of MERS-CoV Receptor Binding Domain with Heparin.Viruses. 2024 Feb 2;16(2):237. doi: 10.3390/v16020237. Viruses. 2024. PMID: 38400013 Free PMC article.
-
A stapled lipopeptide platform for preventing and treating highly pathogenic viruses of pandemic potential.Nat Commun. 2024 Jan 4;15(1):274. doi: 10.1038/s41467-023-44361-1. Nat Commun. 2024. PMID: 38177138 Free PMC article.
-
The role of angiotensin-converting enzyme 2 (ACE2) genetic variations in COVID-19 infection: a literature review.Egypt J Med Hum Genet. 2022;23(1):97. doi: 10.1186/s43042-022-00309-6. Epub 2022 May 28. Egypt J Med Hum Genet. 2022. PMID: 37521836 Free PMC article. Review.
-
Targeting the SARS-CoV-2 HR1 with Small Molecules as Inhibitors of the Fusion Process.Int J Mol Sci. 2022 Sep 3;23(17):10067. doi: 10.3390/ijms231710067. Int J Mol Sci. 2022. PMID: 36077465 Free PMC article.
-
Computational studies on potential new anti-Covid-19 agents with a multi-target mode of action.J King Saud Univ Sci. 2022 Jul;34(5):102086. doi: 10.1016/j.jksus.2022.102086. Epub 2022 May 13. J King Saud Univ Sci. 2022. PMID: 35582633 Free PMC article.
References
-
- Poutanen S.M., Low D.E., Henry B., Finkelstein S., Rose D., Green K., Tellier R., Draker R., Adachi D., Ayers M., Chan A.K., Skowronski D.M., Salit I., Simor A.E., Slutsky A.S., Doyle P.W., Krajden M., Petric M., Brunham R.C., McGeer A.J. N. Engl. J. Med. 2003;348:1995–2005. - PubMed
-
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A., Berger A., Burguiere A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Muller S., Rickerts V., Sturmer M., Vieth S., Klenk H.D., Osterhaus A.D., Schmitz H., Doerr H.W. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Eickmann M., Becker S., Klenk H.D., Doerr H.W., Stadler K., Censini S., Guidotti S., Masignani V., Scarselli M., Mora M., Donati C., Han J.H., Song H.C., Abrignani S., Covacci A., Rappuoli R. Science. 2003;302:1504–1505. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

