Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably?
- PMID: 15345057
- PMCID: PMC516451
- DOI: 10.1186/1742-4690-1-23
Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably?
Abstract
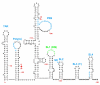
During virus assembly, all retroviruses specifically encapsidate two copies of full-length viral genomic RNA in the form of a non-covalently linked RNA dimer. The absolute conservation of this unique genome structure within the Retroviridae family is strong evidence that a dimerized genome is of critical importance to the viral life cycle. An obvious hypothesis is that retroviruses have evolved to preferentially package two copies of genomic RNA, and that dimerization ensures the proper packaging specificity for such a genome. However, this implies that dimerization must be a prerequisite for genome encapsidation, a notion that has been debated for many years. In this article, we review retroviral RNA dimerization and packaging, highlighting the research that has attempted to dissect the intricate relationship between these two processes in the context of HIV-1, and discuss the therapeutic potential of these putative antiretroviral targets.
Figures
Similar articles
-
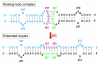
Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248-271 are dispensable for dimer formation.Biochemistry. 1996 Feb 6;35(5):1589-98. doi: 10.1021/bi951838f. Biochemistry. 1996. PMID: 8634290
-
Cis elements and trans-acting factors involved in the RNA dimerization of the human immunodeficiency virus HIV-1.J Mol Biol. 1990 Dec 5;216(3):689-99. doi: 10.1016/0022-2836(90)90392-Y. J Mol Biol. 1990. PMID: 2124274
-
Role of distal zinc finger of nucleocapsid protein in genomic RNA dimerization of human immunodeficiency virus type 1; no role for the palindrome crowning the R-U5 hairpin.Virology. 2001 Mar 1;281(1):109-16. doi: 10.1006/viro.2000.0778. Virology. 2001. PMID: 11222101
-
How retroviruses select their genomes.Nat Rev Microbiol. 2005 Aug;3(8):643-55. doi: 10.1038/nrmicro1210. Nat Rev Microbiol. 2005. PMID: 16064056 Review.
-
Human Retrovirus Genomic RNA Packaging.Viruses. 2022 May 19;14(5):1094. doi: 10.3390/v14051094. Viruses. 2022. PMID: 35632835 Free PMC article. Review.
Cited by
-
Inhibition of 5'-UTR RNA conformational switching in HIV-1 using antisense PNAs.PLoS One. 2012;7(11):e49310. doi: 10.1371/journal.pone.0049310. Epub 2012 Nov 12. PLoS One. 2012. PMID: 23152893 Free PMC article.
-
Effects of nucleic acid local structure and magnesium ions on minus-strand transfer mediated by the nucleic acid chaperone activity of HIV-1 nucleocapsid protein.Nucleic Acids Res. 2007;35(12):3974-87. doi: 10.1093/nar/gkm375. Epub 2007 Jun 6. Nucleic Acids Res. 2007. PMID: 17553835 Free PMC article.
-
Analysis of the contribution of reverse transcriptase and integrase proteins to retroviral RNA dimer conformation.J Virol. 2005 May;79(10):6338-48. doi: 10.1128/JVI.79.10.6338-6348.2005. J Virol. 2005. PMID: 15858017 Free PMC article.
-
Selective packaging of HIV-1 RNA genome is guided by the stability of 5' untranslated region polyA stem.Proc Natl Acad Sci U S A. 2021 Dec 14;118(50):e2114494118. doi: 10.1073/pnas.2114494118. Proc Natl Acad Sci U S A. 2021. PMID: 34873042 Free PMC article.
-
Structural determinants and mechanism of HIV-1 genome packaging.J Mol Biol. 2011 Jul 22;410(4):609-33. doi: 10.1016/j.jmb.2011.04.029. J Mol Biol. 2011. PMID: 21762803 Free PMC article. Review.
References
-
- Greatorex J, Lever A. Retroviral RNA dimer linkage. J Gen Virol. 1998;79:2877–2882. - PubMed
-
- Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog Nucleic Acid Res Mol Biol. 1996;54:1–34. - PubMed
-
- Lever AM, Richardson JH, Harrison GP. Retroviral RNA packaging. Biochem Soc Trans. 1991;19:963–966. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

