Genetic relationships and evolution of genotypes of yellow fever virus and other members of the yellow fever virus group within the Flavivirus genus based on the 3' noncoding region
- PMID: 15331698
- PMCID: PMC515011
- DOI: 10.1128/JVI.78.18.9652-9665.2004
Genetic relationships and evolution of genotypes of yellow fever virus and other members of the yellow fever virus group within the Flavivirus genus based on the 3' noncoding region
Abstract
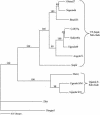
Genetic relationships among flaviviruses within the yellow fever (YF) virus genetic group were investigated by comparing nucleotide sequences of the 3' noncoding region (3'NCR). Size heterogeneity was observed between members and even among strains of the same viral species. Size variation between YF strains was due to duplications and/or deletions of repeated nucleotide sequence elements (RYF). West African genotypes had three copies of the RYF (RYF1, RYF2, and RYF3); the Angola and the East and Central African genotypes had two copies (RYF1 and RYF3); and South American genotypes had only a single copy (RYF3). Nucleotide sequence analyses suggest a deletion within the 3'NCR of South American genotypes, including RYF1 and RYF2. Based on studies with the French neurotropic vaccine strain, passage of a YF virus strain in cell culture can result in deletion of RYF1 and RYF2. Taken together, these observations suggest that South American genotypes of YF virus evolved from West African genotypes and that the South American genotypes lost RYF1 and RYF2, possibly in a single event. Repeated sequence elements were found within the 3'NCR of other members of the YF virus genetic group, suggesting that it is probably characteristic for members of the YF virus genetic group. A core sequence of 15 nucleotides, containing two stem-loops, was found within the 3'NCR of all members of the YF genetic group and may represent the progenitor repeat sequence. Secondary structure predictions of the 3'NCR showed very similar structures for viruses that were closely related phylogenetically.
Copyright 2004 American Society for Microbiology
Figures
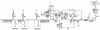



Similar articles
-
Size heterogeneity in the 3' noncoding region of South American isolates of yellow fever virus.J Virol. 2005 Mar;79(6):3807-21. doi: 10.1128/JVI.79.6.3807-3821.2005. J Virol. 2005. PMID: 15731274 Free PMC article.
-
Genetic variation in yellow fever virus: duplication in the 3' noncoding region of strains from Africa.Virology. 1996 Nov 15;225(2):274-81. doi: 10.1006/viro.1996.0601. Virology. 1996. PMID: 8918913
-
Phylogenetic and evolutionary relationships among yellow fever virus isolates in Africa.J Virol. 2001 Aug;75(15):6999-7008. doi: 10.1128/JVI.75.15.6999-7008.2001. J Virol. 2001. PMID: 11435580 Free PMC article.
-
Yellow fever: a disease that has yet to be conquered.Annu Rev Entomol. 2007;52:209-29. doi: 10.1146/annurev.ento.52.110405.091454. Annu Rev Entomol. 2007. PMID: 16913829 Review.
-
Phylogeographic reconstruction of African yellow fever virus isolates indicates recent simultaneous dispersal into east and west Africa.PLoS Negl Trop Dis. 2013;7(3):e1910. doi: 10.1371/journal.pntd.0001910. Epub 2013 Mar 14. PLoS Negl Trop Dis. 2013. PMID: 23516640 Free PMC article. Review.
Cited by
-
Phylogenomic analysis unravels evolution of yellow fever virus within hosts.PLoS Negl Trop Dis. 2018 Sep 6;12(9):e0006738. doi: 10.1371/journal.pntd.0006738. eCollection 2018 Sep. PLoS Negl Trop Dis. 2018. PMID: 30188905 Free PMC article.
-
Functions of the 3' and 5' genome RNA regions of members of the genus Flavivirus.Virus Res. 2015 Aug 3;206:108-19. doi: 10.1016/j.virusres.2015.02.006. Epub 2015 Feb 13. Virus Res. 2015. PMID: 25683510 Free PMC article. Review.
-
What Does the Future Hold for Yellow Fever Virus? (II).Genes (Basel). 2018 Aug 21;9(9):425. doi: 10.3390/genes9090425. Genes (Basel). 2018. PMID: 30134625 Free PMC article. Review.
-
Construction and characterization of recombinant flaviviruses bearing insertions between E and NS1 genes.Virol J. 2007 Oct 30;4:115. doi: 10.1186/1743-422X-4-115. Virol J. 2007. PMID: 17971212 Free PMC article.
-
Yellow fever in Africa and the Americas: a historical and epidemiological perspective.J Venom Anim Toxins Incl Trop Dis. 2018 Aug 25;24:20. doi: 10.1186/s40409-018-0162-y. eCollection 2018. J Venom Anim Toxins Incl Trop Dis. 2018. PMID: 30158957 Free PMC article. Review.
References
-
- Batista, W. C., S. Kashima, C. Marques, and L. T. M. Figueiredo. 2001. Phylogenetic analysis of Brazilian flaviviruses using nucleotide sequences of parts of NS5 gene and 3′ non-coding regions. Virus Res. 75:35-42. - PubMed
-
- Beasley, D. W. C., M. T. Suderman, M. R. Holbrook, and A. D. T. Barrett. 2001. Nucleotide sequencing and serological evidence that the recently recognized deer tick virus is a genotype of Powassan virus. Virus Res. 79:81-89. - PubMed
-
- Billoir, F., R. de Chesse, H. Tolou de Micco, E. A. Gould, and X. de Lamballerie. 2000. Phylogeny of the genus Flavivirus using complete coding sequences of arthropod-borne viruses and viruses with no known vector. J. Gen. Virol. 81:781-790. - PubMed
-
- Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huikman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261-1268. - PubMed
-
- Calisher, C. H., N. Karabatsos, J. M. Dalrymple, R. E. Shope, J. S. Porterfield, E. G. Westway, and W. E. Brandt. 1989. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 70:37-43. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

