A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication
- PMID: 15329670
- PMCID: PMC517609
- DOI: 10.1038/sj.emboj.7600369
A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication
Abstract
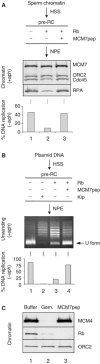
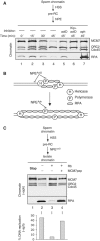
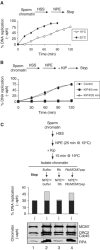
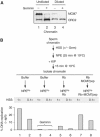
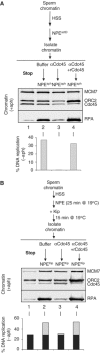
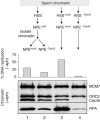
In vertebrates, MCM2-7 and Cdc45 are required for DNA replication initiation, but it is unknown whether they are also required for elongation, as in yeast. Moreover, although MCM2-7 is a prime candidate for the eukaryotic replicative DNA helicase, a demonstration that MCM2-7 unwinds DNA during replication is lacking. Here, we use Xenopus egg extracts to investigate the roles of MCM7 and Cdc45 in DNA replication. A fragment of the retinoblastoma protein, Rb(1-400), was used to neutralize MCM7, and antibodies were used to neutralize Cdc45. When added immediately after origin unwinding, or after significant DNA synthesis, both inhibitors blocked further DNA replication, indicating that MCM7 and Cdc45 are required throughout replication elongation in vertebrates. We next exploited the fact that inhibition of DNA polymerase by aphidicolin causes extensive chromosome unwinding, likely due to uncoupling of the replicative DNA helicase. Strikingly, Rb(1-400) and Cdc45 antibodies both abolished unwinding by the uncoupled helicase. These results provide new support for the model that MCM2-7 is the replicative DNA helicase, and they indicate that Cdc45 functions as a helicase co-factor.
Figures
Similar articles
-
The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication.J Cell Biol. 2002 Nov 25;159(4):541-7. doi: 10.1083/jcb.200207090. Epub 2002 Nov 18. J Cell Biol. 2002. PMID: 12438414 Free PMC article.
-
Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication.Mol Cell. 2006 Feb 17;21(4):581-7. doi: 10.1016/j.molcel.2006.01.030. Mol Cell. 2006. PMID: 16483939
-
Mcm10 coordinates the timely assembly and activation of the replication fork helicase.Nucleic Acids Res. 2016 Jan 8;44(1):315-29. doi: 10.1093/nar/gkv1260. Epub 2015 Nov 17. Nucleic Acids Res. 2016. PMID: 26582917 Free PMC article.
-
The eukaryotic Mcm2-7 replicative helicase.Subcell Biochem. 2012;62:113-34. doi: 10.1007/978-94-007-4572-8_7. Subcell Biochem. 2012. PMID: 22918583 Review.
-
Activation of the replicative DNA helicase: breaking up is hard to do.Curr Opin Cell Biol. 2012 Jun;24(3):423-30. doi: 10.1016/j.ceb.2012.01.011. Epub 2012 Mar 16. Curr Opin Cell Biol. 2012. PMID: 22424671 Review.
Cited by
-
Cell cycle arrest by transforming growth factor beta1 near G1/S is mediated by acute abrogation of prereplication complex activation involving an Rb-MCM interaction.Mol Cell Biol. 2010 Feb;30(3):845-56. doi: 10.1128/MCB.01152-09. Epub 2009 Nov 30. Mol Cell Biol. 2010. PMID: 19948884 Free PMC article.
-
Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins.Proc Natl Acad Sci U S A. 2009 Sep 15;106(37):15628-32. doi: 10.1073/pnas.0908039106. Epub 2009 Sep 8. Proc Natl Acad Sci U S A. 2009. PMID: 19805216 Free PMC article.
-
Characterization of functional domains in human Claspin.Cell Cycle. 2011 May 15;10(10):1599-606. doi: 10.4161/cc.10.10.15562. Epub 2011 May 15. Cell Cycle. 2011. PMID: 21478680 Free PMC article.
-
The Human Replicative Helicase, the CMG Complex, as a Target for Anti-cancer Therapy.Front Mol Biosci. 2018 Mar 29;5:26. doi: 10.3389/fmolb.2018.00026. eCollection 2018. Front Mol Biosci. 2018. PMID: 29651420 Free PMC article. Review.
-
ATR limits Rad18-mediated PCNA monoubiquitination to preserve replication fork and telomerase-independent telomere stability.EMBO J. 2024 Apr;43(7):1301-1324. doi: 10.1038/s44318-024-00066-9. Epub 2024 Mar 11. EMBO J. 2024. PMID: 38467834 Free PMC article.
References
-
- Aparicio OM, Weinstein DM, Bell SP (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69 - PubMed
-
- Avni D, Yang H, Martelli F, Hofmann F, ElShamy WM, Ganesan S, Scully R, Livingston DM (2003) Active localization of the retinoblastoma protein in chromatin and its response to S phase DNA damage. Mol Cell 12: 735–746 - PubMed
-
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 - PubMed
-
- Blow JJ, Laskey RA (1986) Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell 47: 577–587 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

