BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development
- PMID: 15317862
- PMCID: PMC6729778
- DOI: 10.1523/JNEUROSCI.1739-04.2004
BDNF regulates the translation of a select group of mRNAs by a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway during neuronal development
Abstract
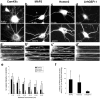
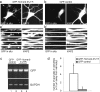
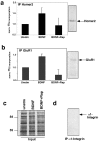
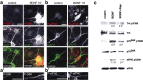
Local regulation of mRNA translation plays an important role in axon guidance, synaptic development, and neuronal plasticity. Little is known, however, regarding the mechanisms that control translation in neurons, and only a few mRNAs have been identified that are locally translated within axon and dendrites. Using Affymetrix gene arrays to identify mRNAs that are newly associated with polysomes after exposure to BDNF, we identified subsets of mRNAs for which translation is enhanced in neurons at different developmental stages. In mature neurons, many of these mRNAs encode proteins that are known to function at synapses, including CamKIIalpha, NMDA receptor subunits, and the postsynaptic density (PSD) scaffolding protein Homer2. BDNF regulates the translation of Homer2 locally in the synaptodendritic compartment by activating translational initiation via a mammalian target of rapamycin-phosphatidylinositol 3-kinase-dependent pathway. These findings suggest that BDNF likely regulates synaptic function by inducing the local synthesis of numerous synaptic proteins. The local translation of the cytoskeleton-associated protein Homer2 in particular might have important implications for growth cone dynamics and dendritic spine development.
Figures
Similar articles
-
BDNF-induced local protein synthesis and synaptic plasticity.Neuropharmacology. 2014 Jan;76 Pt C:639-56. doi: 10.1016/j.neuropharm.2013.04.005. Epub 2013 Apr 16. Neuropharmacology. 2014. PMID: 23602987 Review.
-
A rapamycin-sensitive signaling pathway contributes to long-term synaptic plasticity in the hippocampus.Proc Natl Acad Sci U S A. 2002 Jan 8;99(1):467-72. doi: 10.1073/pnas.012605299. Epub 2001 Dec 26. Proc Natl Acad Sci U S A. 2002. PMID: 11756682 Free PMC article.
-
Insulin stimulates postsynaptic density-95 protein translation via the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway.J Biol Chem. 2005 May 6;280(18):18543-50. doi: 10.1074/jbc.M414112200. Epub 2005 Mar 8. J Biol Chem. 2005. PMID: 15755733
-
Enhancement of translation elongation in neurons by brain-derived neurotrophic factor: implications for mammalian target of rapamycin signaling.J Neurochem. 2005 Dec;95(5):1438-45. doi: 10.1111/j.1471-4159.2005.03466.x. Epub 2005 Sep 20. J Neurochem. 2005. PMID: 16171514
-
Regulation of local translation at the synapse by BDNF.Prog Neurobiol. 2010 Dec;92(4):505-16. doi: 10.1016/j.pneurobio.2010.08.004. Epub 2010 Aug 14. Prog Neurobiol. 2010. PMID: 20713125 Review.
Cited by
-
Dual regulation of miRNA biogenesis generates target specificity in neurotrophin-induced protein synthesis.Cell. 2012 Mar 2;148(5):933-46. doi: 10.1016/j.cell.2012.01.036. Cell. 2012. PMID: 22385959 Free PMC article.
-
AMPA receptor trafficking in homeostatic synaptic plasticity: functional molecules and signaling cascades.Neural Plast. 2012;2012:825364. doi: 10.1155/2012/825364. Epub 2012 May 13. Neural Plast. 2012. PMID: 22655210 Free PMC article. Review.
-
Developing and applying the adverse outcome pathway concept for understanding and predicting neurotoxicity.Neurotoxicology. 2017 Mar;59:240-255. doi: 10.1016/j.neuro.2016.05.010. Epub 2016 May 17. Neurotoxicology. 2017. PMID: 27212452 Free PMC article. Review.
-
RNA trafficking and local protein synthesis in dendrites: an overview.J Neurosci. 2006 Jul 5;26(27):7131-4. doi: 10.1523/JNEUROSCI.1801-06.2006. J Neurosci. 2006. PMID: 16822966 Free PMC article. Review.
-
Non coding RNA and brain.BMC Neurosci. 2006 Oct 30;7 Suppl 1(Suppl 1):S5. doi: 10.1186/1471-2202-7-S1-S5. BMC Neurosci. 2006. PMID: 17118159 Free PMC article. Review.
References
-
- Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM (2001) Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 30: 489-502. - PubMed
-
- Aizawa H, Wakatsuki S, Ishii A, Moriyama K, Sasaki Y, Ohashi K, Sekine-Aizawa Y, Sehara-Fujisawa A, Mizuno K, Goshima Y, Yahara I (2001) Phosphorylation of cofilin by LIM-kinase is necessary for semaphorin 3A-induced growth cone collapse. Nat Neurosci 4: 367-373. - PubMed
-
- Bonni A, Greenberg ME (1997) Neurotrophin regulation of gene expression. Can J Neurol Sci 24: 272-283. - PubMed
-
- Brittis PA, Lu Q, Flanagan JG (2002) Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell 110: 223-235. - PubMed
-
- Campbell DS, Holt CE (2001) Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron 32: 1013-1026. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

