Vif is an auxiliary factor of the HIV-1 reverse transcriptase and facilitates abasic site bypass
- PMID: 15315477
- PMCID: PMC1133740
- DOI: 10.1042/BJ20040914
Vif is an auxiliary factor of the HIV-1 reverse transcriptase and facilitates abasic site bypass
Abstract
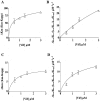
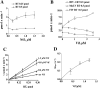
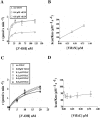
The HIV-1 accessory protein Vif was found to modulate the RNA- and DNA-dependent DNA synthesis activity of the viral RT (reverse transcriptase) in two ways: (i) it stimulated the binding of the viral RT to the primer by increasing the association rate kcat/K(m) and by decreasing the thermodynamic barrier DeltaH([ES]) for complex formation, and (ii) it increased the polymerization rate of HIV-1 RT. A Vif mutant lacking the final 56 amino acids at the C-terminus failed to stimulate the viral RT. On the other hand, another Vif mutant lacking the first 43 amino acids at the N-terminus, which are involved in RNA binding and interaction with the viral protease, was able to stimulate RT activity. In addition, Vif was found to promote the bypass of an abasic site by HIV-1 RT.
Figures


Similar articles
-
A drug resistance mutation in the inhibitor binding pocket of human immunodeficiency virus type 1 reverse transcriptase impairs DNA synthesis and RNA degradation.Biochemistry. 1996 Jul 30;35(30):9737-45. doi: 10.1021/bi9600308. Biochemistry. 1996. PMID: 8703945
-
Cooperative and specific binding of Vif to the 5' region of HIV-1 genomic RNA.J Mol Biol. 2005 Nov 18;354(1):55-72. doi: 10.1016/j.jmb.2005.09.025. Epub 2005 Oct 3. J Mol Biol. 2005. PMID: 16236319
-
p53 enhances the fidelity of DNA synthesis by human immunodeficiency virus type 1 reverse transcriptase.Oncogene. 2001 Nov 15;20(52):7635-44. doi: 10.1038/sj.onc.1204956. Oncogene. 2001. PMID: 11753641
-
Interactions between human immunodeficiency virus type 1 reverse transcriptase, tRNA primer, and nucleocapsid protein during reverse transcription.J Hum Virol. 2000 Jan-Feb;3(1):16-26. J Hum Virol. 2000. PMID: 10774803 Review.
-
'Binding, bending and bonding': polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus.Int J Biochem Cell Biol. 2004 Sep;36(9):1752-66. doi: 10.1016/j.biocel.2004.02.016. Int J Biochem Cell Biol. 2004. PMID: 15183342 Review.
Cited by
-
Tumultuous relationship between the human immunodeficiency virus type 1 viral infectivity factor (Vif) and the human APOBEC-3G and APOBEC-3F restriction factors.Microbiol Mol Biol Rev. 2009 Jun;73(2):211-32. doi: 10.1128/MMBR.00040-08. Microbiol Mol Biol Rev. 2009. PMID: 19487726 Free PMC article. Review.
-
Mutational analysis of the HIV-1 auxiliary protein Vif identifies independent domains important for the physical and functional interaction with HIV-1 reverse transcriptase.Nucleic Acids Res. 2009 Jun;37(11):3660-9. doi: 10.1093/nar/gkp226. Epub 2009 Apr 15. Nucleic Acids Res. 2009. PMID: 19369217 Free PMC article.
-
Structural analysis of viral infectivity factor of HIV type 1 and its interaction with A3G, EloC and EloB.PLoS One. 2014 Feb 26;9(2):e89116. doi: 10.1371/journal.pone.0089116. eCollection 2014. PLoS One. 2014. PMID: 24586532 Free PMC article.
-
Vif is a RNA chaperone that could temporally regulate RNA dimerization and the early steps of HIV-1 reverse transcription.Nucleic Acids Res. 2007;35(15):5141-53. doi: 10.1093/nar/gkm542. Epub 2007 Jul 26. Nucleic Acids Res. 2007. PMID: 17660191 Free PMC article.
-
APOBEC3G contributes to HIV-1 variation through sublethal mutagenesis.J Virol. 2010 Jul;84(14):7396-404. doi: 10.1128/JVI.00056-10. Epub 2010 May 12. J Virol. 2010. PMID: 20463080 Free PMC article.
References
-
- Frankel A. D., Young J. A. HIV-1: fifteen proteins and an RNA. Annu. Rev. Biochem. 1998;67:1–25. - PubMed
-
- Gabuzda D. H., Li H., Lawrence K., Vasir B. S., Crawford K., Langhoff E. Essential role of vif in establishing productive HIV-1 infection in peripheral blood T lymphocytes and monocyte/macrophages. J. Acquired Immune Defic. Syndr. 1994;7:908–915. - PubMed
-
- Inubushi R., Adachi A. Cell-dependent function of HIV-1 Vif for virus replication. Int. J. Mol. Med. 1999;3:473–476. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

