Foxa2 regulates multiple pathways of insulin secretion
- PMID: 15314688
- PMCID: PMC503770
- DOI: 10.1172/JCI21149
Foxa2 regulates multiple pathways of insulin secretion
Abstract
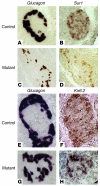
The regulation of insulin secretion by pancreatic beta cells is perturbed in several diseases, including adult-onset (type 2) diabetes and persistent hyperinsulinemic hypoglycemia of infancy (PHHI). The first mouse model for PHHI has a conditional deletion of the gene encoding the winged-helix transcription factor Foxa2 (Forkhead box a2, formerly Hepatocyte nuclear factor 3beta) in pancreatic beta cells. Using isolated islets, we found that Foxa2 deficiency resulted in excessive insulin release in response to amino acids and complete loss of glucose-stimulated insulin secretion. Most PHHI cases are associated with mutations in SUR1 (Sulfonylurea receptor 1) or KIR6.2 (Inward rectifier K(+) channel member 6.2), which encode the subunits of the ATP-sensitive K(+) channel, and RNA in situ hybridization of mutant mouse islets revealed that expression of both genes is Foxa2 dependent. We utilized expression profiling to identify additional targets of Foxa2. Strikingly, one of these genes, Hadhsc, encodes short-chain L-3-hydroxyacyl-coenzyme A dehydrogenase, deficiency of which has been shown to cause PHHI in humans. Hadhsc is a direct target of Foxa2, as demonstrated by cotransfection as well as in vivo chromatin immunoprecipitation experiments using isolated islets. Thus, we have established Foxa2 as an essential activator of genes that function in multiple pathways governing insulin secretion.
Figures




Similar articles
-
Engineering a glucose-responsive human insulin-secreting cell line from islets of Langerhans isolated from a patient with persistent hyperinsulinemic hypoglycemia of infancy.J Biol Chem. 1999 Nov 26;274(48):34059-66. doi: 10.1074/jbc.274.48.34059. J Biol Chem. 1999. PMID: 10567373
-
Hepatocyte nuclear factor-4alpha is essential for glucose-stimulated insulin secretion by pancreatic beta-cells.J Biol Chem. 2006 Feb 24;281(8):5246-57. doi: 10.1074/jbc.M507496200. Epub 2005 Dec 23. J Biol Chem. 2006. PMID: 16377800
-
Cholinergic regulation of fuel-induced hormone secretion and respiration of SUR1-/- mouse islets.Am J Physiol Endocrinol Metab. 2006 Sep;291(3):E525-35. doi: 10.1152/ajpendo.00579.2005. Epub 2006 Apr 25. Am J Physiol Endocrinol Metab. 2006. PMID: 16638820
-
Winged-helix transcription factors and pancreatic development.Clin Sci (Lond). 2005 Mar;108(3):195-204. doi: 10.1042/CS20040309. Clin Sci (Lond). 2005. PMID: 15631623 Review.
-
Roles of KATP channels as metabolic sensors in acute metabolic changes.J Mol Cell Cardiol. 2005 Jun;38(6):917-25. doi: 10.1016/j.yjmcc.2004.11.019. Epub 2005 Feb 5. J Mol Cell Cardiol. 2005. PMID: 15910876 Review.
Cited by
-
Organogenesis and functional genomics of the endocrine pancreas.Cell Mol Life Sci. 2012 Jul;69(13):2109-23. doi: 10.1007/s00018-011-0915-z. Epub 2012 Jan 13. Cell Mol Life Sci. 2012. PMID: 22241333 Free PMC article. Review.
-
Hepatocyte-specific ablation of Foxa2 alters bile acid homeostasis and results in endoplasmic reticulum stress.Nat Med. 2008 Aug;14(8):828-36. doi: 10.1038/nm.1853. Epub 2008 Jul 27. Nat Med. 2008. PMID: 18660816 Free PMC article.
-
Mechanisms of the amplifying pathway of insulin secretion in the β cell.Pharmacol Ther. 2017 Nov;179:17-30. doi: 10.1016/j.pharmthera.2017.05.003. Epub 2017 May 18. Pharmacol Ther. 2017. PMID: 28527919 Free PMC article. Review.
-
miR-92a promotes hepatocellular carcinoma cells proliferation and invasion by FOXA2 targeting.Iran J Basic Med Sci. 2017 Jul;20(7):783-790. doi: 10.22038/IJBMS.2017.9010. Iran J Basic Med Sci. 2017. PMID: 28852443 Free PMC article.
-
Diabetes in the post-GWAS era.Nat Genet. 2015 Dec;47(12):1373-4. doi: 10.1038/ng.3453. Nat Genet. 2015. PMID: 26620109
References
-
- Ben-Shushan E, Marshak S, Shoshkes M, Cerasi E, Melloul D. A pancreatic beta-cell-specific enhancer in the human PDX-1 gene is regulated by hepatocyte nuclear factor 3beta (HNF-3beta), HNF-1alpha, and SPs transcription factors. J. Biol. Chem. 2001;276:17533–17540. - PubMed
-
- Weinstein DC, et al. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell. 1994;78:575–588. - PubMed
-
- Ang SL, Rossant J. HNF-3 beta is essential for node and notochord formation in mouse development. Cell. 1994;78:561–574. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

