GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination
- PMID: 15314182
- PMCID: PMC506980
- DOI: 10.1128/MCB.24.17.7769-7778.2004
GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination
Abstract
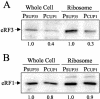
Translation termination in eukaryotes is mediated by two release factors, eRF1 and eRF3. eRF1 recognizes each of the three stop codons (UAG, UAA, and UGA) and facilitates release of the nascent polypeptide chain. eRF3 is a GTPase that stimulates the translation termination process by a poorly characterized mechanism. In this study, we examined the functional importance of GTP hydrolysis by eRF3 in Saccharomyces cerevisiae. We found that mutations that reduced the rate of GTP hydrolysis also reduced the efficiency of translation termination at some termination signals but not others. As much as a 17-fold decrease in the termination efficiency was observed at some tetranucleotide termination signals (characterized by the stop codon and the first following nucleotide), while no effect was observed at other termination signals. To determine whether this stop signal-dependent decrease in the efficiency of translation termination was due to a defect in either eRF1 or eRF3 recycling, we reduced the level of eRF1 or eRF3 in cells by expressing them individually from the CUP1 promoter. We found that the limitation of either factor resulted in a general decrease in the efficiency of translation termination rather than a decrease at a subset of termination signals as observed with the eRF3 GTPase mutants. We also found that overproduction of eRF1 was unable to increase the efficiency of translation termination at any termination signals. Together, these results suggest that the GTPase activity of eRF3 is required to couple the recognition of translation termination signals by eRF1 to efficient polypeptide chain release.
Copyright 2004 American Society for Microbiology
Figures


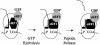
Similar articles
-
N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination.BMC Mol Biol. 2006 Oct 11;7:34. doi: 10.1186/1471-2199-7-34. BMC Mol Biol. 2006. PMID: 17034622 Free PMC article.
-
Eukaryotic polypeptide chain release factor eRF3 is an eRF1- and ribosome-dependent guanosine triphosphatase.RNA. 1996 Apr;2(4):334-41. RNA. 1996. PMID: 8634914 Free PMC article.
-
Ribosome-bound Pub1 modulates stop codon decoding during translation termination in yeast.FEBS J. 2017 Jun;284(12):1914-1930. doi: 10.1111/febs.14099. Epub 2017 May 29. FEBS J. 2017. PMID: 28467675
-
Translation termination and its regulation in eukaryotes: recent insights provided by studies in yeast.Biochemistry (Mosc). 1999 Dec;64(12):1360-6. Biochemistry (Mosc). 1999. PMID: 10648959 Review.
-
Termination of translation in eukaryotes.Biochem Cell Biol. 1995 Nov-Dec;73(11-12):1079-86. doi: 10.1139/o95-116. Biochem Cell Biol. 1995. PMID: 8722024 Review.
Cited by
-
Structural basis for translation termination by archaeal RF1 and GTP-bound EF1α complex.Nucleic Acids Res. 2012 Oct;40(18):9319-28. doi: 10.1093/nar/gks660. Epub 2012 Jul 5. Nucleic Acids Res. 2012. PMID: 22772989 Free PMC article.
-
Suppression of premature termination codons as a therapeutic approach.Crit Rev Biochem Mol Biol. 2012 Sep;47(5):444-63. doi: 10.3109/10409238.2012.694846. Epub 2012 Jun 7. Crit Rev Biochem Mol Biol. 2012. PMID: 22672057 Free PMC article. Review.
-
A single amino acid change of translation termination factor eRF1 switches between bipotent and omnipotent stop-codon specificity.Nucleic Acids Res. 2011 Jan;39(2):599-608. doi: 10.1093/nar/gkq759. Epub 2010 Sep 21. Nucleic Acids Res. 2011. PMID: 20860996 Free PMC article.
-
Eukaryotic ribosomal RNA determinants of aminoglycoside resistance and their role in translational fidelity.RNA. 2008 Jan;14(1):148-57. doi: 10.1261/rna.805208. Epub 2007 Nov 14. RNA. 2008. PMID: 18003936 Free PMC article.
-
Translation Phases in Eukaryotes.Methods Mol Biol. 2022;2533:217-228. doi: 10.1007/978-1-0716-2501-9_13. Methods Mol Biol. 2022. PMID: 35796991 Free PMC article. Review.
References
-
- Alberts, B., A. Johnson, J. Lewis, M. Raff, K. Roberts, and P. Walter. 2002. Molecular Biology of the Cell, 4th ed., p. 616. Garland Science, New York, N.Y.
-
- Anborgh, P. H., R. H. Cool, F. Gumusel, K. Harmark, E. Jacquet, A. Weijland, M. Y. Mistou, and A. Parmeggiani. 1991. Structure-function relationships of elongation factor Tu as studied by mutagenesis. Biochimie 73:1051-1059. - PubMed
-
- Bonetti, B., L. Fu, J. Moon, and D. M. Bedwell. 1995. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 251:334-345. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
