RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia
- PMID: 15314149
- PMCID: PMC506981
- DOI: 10.1128/MCB.24.17.7370-7379.2004
RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia
Abstract
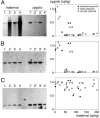
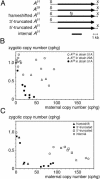
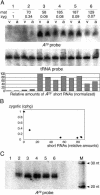
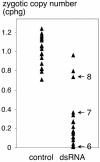
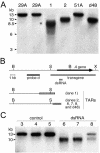
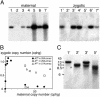
The germ line genome of ciliates is extensively rearranged during development of the somatic macronucleus. Numerous sequences are eliminated, while others are amplified to a high ploidy level. In the Paramecium aurelia group of species, transformation of the maternal macronucleus with transgenes at high copy numbers can induce the deletion of homologous genes in sexual progeny, when a new macronucleus develops from the wild-type germ line. We show that this trans-nuclear effect correlates with homology-dependent silencing of maternal genes before autogamy and with the accumulation of approximately 22- to 23-nucleotide (nt) RNA molecules. The same effects are induced by feeding cells before meiosis with bacteria containing double-stranded RNA, suggesting that small interfering RNA-like molecules can target deletions. Furthermore, experimentally induced macronuclear deletions are spontaneously reproduced in subsequent sexual generations, and reintroduction of the missing gene into the variant macronucleus restores developmental amplification in sexual progeny. We discuss the possible roles of the approximately 22- to 23-nt RNAs in the targeting of deletions and the implications for the RNA-mediated genome-scanning process that is thought to determine developmentally regulated rearrangements in ciliates.
Copyright 2004 American Society for Microbiology
Figures

Similar articles
-
Nowa1p and Nowa2p: novel putative RNA binding proteins involved in trans-nuclear crosstalk in Paramecium tetraurelia.Curr Biol. 2005 Sep 20;15(18):1616-28. doi: 10.1016/j.cub.2005.07.033. Curr Biol. 2005. PMID: 16169483
-
Non-Mendelian inheritance and homology-dependent effects in ciliates.Adv Genet. 2002;46:305-37. doi: 10.1016/s0065-2660(02)46011-7. Adv Genet. 2002. PMID: 11931229 Review.
-
Large-scale genome remodelling by the developmentally programmed elimination of germ line sequences in the ciliate Paramecium.Res Microbiol. 2004 Jun;155(5):399-408. doi: 10.1016/j.resmic.2004.01.017. Res Microbiol. 2004. PMID: 15207872 Review.
-
Epigenetic regulation of programmed genomic rearrangements in Paramecium aurelia.J Eukaryot Microbiol. 1996 Nov-Dec;43(6):453-61. doi: 10.1111/j.1550-7408.1996.tb04504.x. J Eukaryot Microbiol. 1996. PMID: 8976603 Review.
-
Programmed DNA deletion as an RNA-guided system of genome defense.Science. 2003 Jun 6;300(5625):1581-4. doi: 10.1126/science.1084737. Science. 2003. PMID: 12791996
Cited by
-
Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway.Mol Cell Biol. 2006 Dec;26(23):8731-42. doi: 10.1128/MCB.01430-06. Epub 2006 Sep 25. Mol Cell Biol. 2006. PMID: 17000759 Free PMC article.
-
snRNA and heterochromatin formation are involved in DNA excision during macronuclear development in stichotrichous ciliates.Eukaryot Cell. 2005 Nov;4(11):1934-41. doi: 10.1128/EC.4.11.1934-1941.2005. Eukaryot Cell. 2005. PMID: 16278460 Free PMC article.
-
Six domesticated PiggyBac transposases together carry out programmed DNA elimination in Paramecium.Elife. 2018 Sep 18;7:e37927. doi: 10.7554/eLife.37927. Elife. 2018. PMID: 30223944 Free PMC article.
-
The SUMO pathway is developmentally regulated and required for programmed DNA elimination in Paramecium tetraurelia.Eukaryot Cell. 2006 May;5(5):806-15. doi: 10.1128/EC.5.5.806-815.2006. Eukaryot Cell. 2006. PMID: 16682458 Free PMC article.
-
Developmentally programmed DNA splicing in Paramecium reveals short-distance crosstalk between DNA cleavage sites.Nucleic Acids Res. 2008 Jun;36(10):3244-51. doi: 10.1093/nar/gkn154. Epub 2008 Apr 17. Nucleic Acids Res. 2008. PMID: 18420657 Free PMC article.
References
-
- Caron, F., and E. Meyer. 1989. Molecular basis of surface antigen variation in paramecia. Annu. Rev. Microbiol. 43:23-42. - PubMed
-
- Cerutti, H. 2003. RNA interference: traveling in the cell and gaining functions? Trends Genet. 19:39-46. - PubMed
-
- Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
