An efficient one-step site-directed and site-saturation mutagenesis protocol
- PMID: 15304544
- PMCID: PMC514394
- DOI: 10.1093/nar/gnh110
An efficient one-step site-directed and site-saturation mutagenesis protocol
Abstract
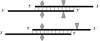
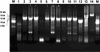
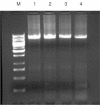
We have developed a new primer design method based on the QuickChange site-directed mutagenesis protocol, which significantly improves the PCR amplification efficiency. This design method minimizes primer dimerization and ensures the priority of primer-template annealing over primer self-pairing during the PCR. Several different multiple mutations (up to 7 bases) were successfully performed with this partial overlapping primer design in a variety of vectors ranging from 4 to 12 kb in length. In comparison, all attempts failed when using complete-overlapping primer pairs as recommended in the standard QuickChange protocol. Our protocol was further extended to site-saturation mutagenesis by introducing randomized codons. Our data indicated no specific sequence selection during library construction, with the randomized positions resulting in average occurrence of each base in each position. This method should be useful to facilitate the preparation of high-quality site saturation libraries.
Figures
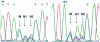
Similar articles
-
A novel megaprimed and ligase-free, PCR-based, site-directed mutagenesis method.Anal Biochem. 2008 Apr 15;375(2):376-8. doi: 10.1016/j.ab.2007.12.013. Epub 2007 Dec 23. Anal Biochem. 2008. PMID: 18198125
-
Polishing the craft of genetic diversity creation in directed evolution.Biotechnol Adv. 2013 Dec;31(8):1707-21. doi: 10.1016/j.biotechadv.2013.08.021. Epub 2013 Sep 6. Biotechnol Adv. 2013. PMID: 24012599 Review.
-
An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol.BMC Biotechnol. 2008 Dec 4;8:91. doi: 10.1186/1472-6750-8-91. BMC Biotechnol. 2008. PMID: 19055817 Free PMC article.
-
A rapid, efficient, and economical inverse polymerase chain reaction-based method for generating a site saturation mutant library.Anal Biochem. 2014 Mar 15;449:90-8. doi: 10.1016/j.ab.2013.12.002. Epub 2013 Dec 9. Anal Biochem. 2014. PMID: 24333246
-
ProxiMAX randomization: a new technology for non-degenerate saturation mutagenesis of contiguous codons.Biochem Soc Trans. 2013 Oct;41(5):1189-94. doi: 10.1042/BST20130123. Biochem Soc Trans. 2013. PMID: 24059507 Free PMC article. Review.
Cited by
-
SRC-3 deficiency prevents atherosclerosis development by decreasing endothelial ICAM-1 expression to attenuate macrophage recruitment.Int J Biol Sci. 2022 Oct 3;18(15):5978-5993. doi: 10.7150/ijbs.74864. eCollection 2022. Int J Biol Sci. 2022. PMID: 36263184 Free PMC article.
-
Structure of plant photosystem I-plastocyanin complex reveals strong hydrophobic interactions.Biochem J. 2021 Jun 25;478(12):2371-2384. doi: 10.1042/BCJ20210267. Biochem J. 2021. PMID: 34085703 Free PMC article.
-
Interaction between basic residues of Epstein-Barr virus EBNA1 protein and cellular chromatin mediates viral plasmid maintenance.J Biol Chem. 2013 Aug 16;288(33):24189-99. doi: 10.1074/jbc.M113.491167. Epub 2013 Jul 8. J Biol Chem. 2013. PMID: 23836915 Free PMC article.
-
Type III-A CRISPR-associated protein Csm6 degrades cyclic hexa-adenylate activator using both CARF and HEPN domains.Nucleic Acids Res. 2020 Sep 18;48(16):9204-9217. doi: 10.1093/nar/gkaa634. Nucleic Acids Res. 2020. PMID: 32766806 Free PMC article.
-
Dynamic regulation of Monascus azaphilones biosynthesis by the binary MrPigE-MrPigF oxidoreductase system.Appl Microbiol Biotechnol. 2022 Nov;106(22):7519-7530. doi: 10.1007/s00253-022-12219-z. Epub 2022 Oct 12. Appl Microbiol Biotechnol. 2022. PMID: 36221033
References
-
- Cormack B. (1994) Introduction of a point mutation by sequential PCR steps. Curr. Protoc. Mol. Biol., 2, 8.5.7–8.5.9.
-
- Aiyar A., Xiang,Y. and Leis,J. (1996) Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol., 57, 177–191. - PubMed
-
- Ishii T.M., Zerr,P., Xia,X.M., Bond,C.T., Maylie,J. and Adelman,J.P. (1998) Site-directed mutagenesis. Methods Enzymol., 293, 53–71. - PubMed
-
- Ling M.M. and Robinson,B.H. (1997) Approaches to DNA mutagenesis: an overview. Anal. Biochem., 254, 157–178. - PubMed
-
- Wang W. and Marcolm,B.A. (1999) Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuickChange™ site-directed mutagenesis. BioTechniques, 26, 680–682. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

