RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa
- PMID: 15302921
- PMCID: PMC514385
- DOI: 10.1093/nar/gkh764
RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa
Abstract
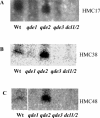
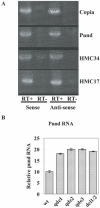
RNA interference (RNAi) can silence genes at the transcriptional level by targeting locus-specific Lys9H3 methylation or at the post-transcriptional level by targeting mRNA degradation. Here we have cloned and sequenced genomic regions methylated in Lys9H3 in Neurospora crassa to test the requirements for components of the RNAi pathway in this modification. We find that 90% of clones map to repeated sequences and relics of transposons that have undergone repeat-induced point mutations (RIP). We find siRNAs derived from transposon relics indicating that the RNAi machinery targets these regions. This is confirmed by the fact that the presence of these siRNAs depends on components of the RNAi pathway such as the RdRP (QDE-1), the putative RecQ helicase (QDE-3) and the two Dicer enzymes. We show that Lys9H3 methylation of RIP sequences is not affected in mutants of the RNAi pathway indicating that the RNAi machinery is not involved in transcriptional gene silencing in Neurospora. We find that RIP regions are transcribed and that the transcript level increases in the mutants of the RNAi pathway. These data suggest that the biological function of the Neurospora RNAi machinery is to control transposon relics and repeated sequences by targeting degradation of transcripts derived from these regions.
Figures
Similar articles
-
Small interfering RNAs that trigger posttranscriptional gene silencing are not required for the histone H3 Lys9 methylation necessary for transgenic tandem repeat stabilization in Neurospora crassa.Mol Cell Biol. 2005 May;25(9):3793-801. doi: 10.1128/MCB.25.9.3793-3801.2005. Mol Cell Biol. 2005. PMID: 15831483 Free PMC article.
-
The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa.Nucleic Acids Res. 2005 Mar 14;33(5):1564-73. doi: 10.1093/nar/gki300. Print 2005. Nucleic Acids Res. 2005. PMID: 15767281 Free PMC article.
-
A double-stranded-RNA response program important for RNA interference efficiency.Mol Cell Biol. 2007 Jun;27(11):3995-4005. doi: 10.1128/MCB.00186-07. Epub 2007 Mar 19. Mol Cell Biol. 2007. PMID: 17371837 Free PMC article.
-
RIP: the evolutionary cost of genome defense.Trends Genet. 2004 Sep;20(9):417-23. doi: 10.1016/j.tig.2004.07.007. Trends Genet. 2004. PMID: 15313550 Review.
-
Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa.Curr Opin Microbiol. 2007 Apr;10(2):199-203. doi: 10.1016/j.mib.2007.03.016. Epub 2007 Mar 28. Curr Opin Microbiol. 2007. PMID: 17395524 Review.
Cited by
-
Advances in Molecular Tools and In Vivo Models for the Study of Human Fungal Pathogenesis.Microorganisms. 2020 May 26;8(6):803. doi: 10.3390/microorganisms8060803. Microorganisms. 2020. PMID: 32466582 Free PMC article. Review.
-
Characterization of Dicer-deficient murine embryonic stem cells.Proc Natl Acad Sci U S A. 2005 Aug 23;102(34):12135-40. doi: 10.1073/pnas.0505479102. Epub 2005 Aug 12. Proc Natl Acad Sci U S A. 2005. PMID: 16099834 Free PMC article.
-
QIP, a protein that converts duplex siRNA into single strands, is required for meiotic silencing by unpaired DNA.Genetics. 2010 Sep;186(1):119-26. doi: 10.1534/genetics.110.118273. Epub 2010 Jun 15. Genetics. 2010. PMID: 20551436 Free PMC article.
-
Mechanism of siRNA production from repetitive DNA.Genes Dev. 2015 Mar 1;29(5):526-37. doi: 10.1101/gad.255828.114. Epub 2015 Feb 17. Genes Dev. 2015. PMID: 25691092 Free PMC article.
-
Targeted gene silencing in the model mushroom Coprinopsis cinerea (Coprinus cinereus) by expression of homologous hairpin RNAs.Eukaryot Cell. 2006 Apr;5(4):732-44. doi: 10.1128/EC.5.4.732-744.2006. Eukaryot Cell. 2006. PMID: 16607020 Free PMC article.
References
-
- Denli A.M. and Hannon,G.J. (2003) RNAi: an ever-growing puzzle. Trends Biochem. Sci., 28, 196–201. - PubMed
-
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. - PubMed
-
- Hammond S.M., Boettcher,S., Caudy,A.A., Kobayashi,R. and Hannon,G.J. (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science, 293, 1146–1150. - PubMed
-
- Volpe T.A., Kidner,C., Hall,I.M., Teng,G., Grewal,S.I. and Martienssen,R.A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science, 297, 1833–1837. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

