Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics
- PMID: 15294028
- PMCID: PMC514697
- DOI: 10.1186/1471-2105-5-104
Design, implementation and evaluation of a practical pseudoknot folding algorithm based on thermodynamics
Abstract
Background: The general problem of RNA secondary structure prediction under the widely used thermodynamic model is known to be NP-complete when the structures considered include arbitrary pseudoknots. For restricted classes of pseudoknots, several polynomial time algorithms have been designed, where the O(n6)time and O(n4) space algorithm by Rivas and Eddy is currently the best available program.
Results: We introduce the class of canonical simple recursive pseudoknots and present an algorithm that requires O(n4) time and O(n2) space to predict the energetically optimal structure of an RNA sequence, possible containing such pseudoknots. Evaluation against a large collection of known pseudoknotted structures shows the adequacy of the canonization approach and our algorithm.
Conclusions: RNA pseudoknots of medium size can now be predicted reliably as well as efficiently by the new algorithm.
Figures


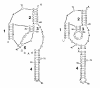
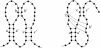
Similar articles
-
A dynamic programming algorithm for RNA structure prediction including pseudoknots.J Mol Biol. 1999 Feb 5;285(5):2053-68. doi: 10.1006/jmbi.1998.2436. J Mol Biol. 1999. PMID: 9925784
-
Thermodynamic heuristics with case-based reasoning: combined insights for RNA pseudoknot secondary structure.J Biomol Struct Dyn. 2011 Aug;29(1):1-26. doi: 10.1080/07391102.2011.10507373. J Biomol Struct Dyn. 2011. PMID: 21696223
-
FlexStem: improving predictions of RNA secondary structures with pseudoknots by reducing the search space.Bioinformatics. 2008 Sep 15;24(18):1994-2001. doi: 10.1093/bioinformatics/btn327. Epub 2008 Jun 27. Bioinformatics. 2008. PMID: 18586700
-
Prediction of RNA secondary structure by free energy minimization.Curr Opin Struct Biol. 2006 Jun;16(3):270-8. doi: 10.1016/j.sbi.2006.05.010. Epub 2006 May 19. Curr Opin Struct Biol. 2006. PMID: 16713706 Review.
-
New developments in structure determination of pseudoknots.Biopolymers. 1998;48(2-3):137-53. doi: 10.1002/(SICI)1097-0282(1998)48:2<137::AID-BIP4>3.0.CO;2-H. Biopolymers. 1998. PMID: 10333742 Review.
Cited by
-
Secondary Structure Predictions for Long RNA Sequences Based on Inversion Excursions and MapReduce.IEEE Int Symp Parallel Distrib Process Workshops Phd Forum. 2013 May;2013:520-529. doi: 10.1109/IPDPSW.2013.109. IEEE Int Symp Parallel Distrib Process Workshops Phd Forum. 2013. PMID: 26023357 Free PMC article.
-
Thermodynamic matchers for the construction of the cuckoo RNA family.RNA Biol. 2015;12(2):197-207. doi: 10.1080/15476286.2015.1017206. RNA Biol. 2015. PMID: 25779873 Free PMC article.
-
Structural analysis of aligned RNAs.Nucleic Acids Res. 2006;34(19):5471-81. doi: 10.1093/nar/gkl692. Epub 2006 Oct 4. Nucleic Acids Res. 2006. PMID: 17020924 Free PMC article.
-
ProbKnot: fast prediction of RNA secondary structure including pseudoknots.RNA. 2010 Oct;16(10):1870-80. doi: 10.1261/rna.2125310. Epub 2010 Aug 10. RNA. 2010. PMID: 20699301 Free PMC article.
-
Advances in RNA structure prediction from sequence: new tools for generating hypotheses about viral RNA structure-function relationships.J Virol. 2009 Jul;83(13):6326-34. doi: 10.1128/JVI.00251-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369331 Free PMC article. Review. No abstract available.
References
-
- Zuker M, Sankoff S. RNA secondary structures and their prediction. Bull Math Biol. 1984;46:591–621.
-
- Hofacker I, Fontana W, Stadler P, Bonhoeffer L, Tacker M, Schuster P. Fast folding and comparison of RNA secondary structures. Monatshefte Chemie. 1994;125:167–188.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

