Crystal structure of human otubain 2
- PMID: 15258613
- PMCID: PMC1299112
- DOI: 10.1038/sj.embor.7400201
Crystal structure of human otubain 2
Abstract
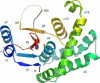
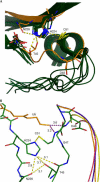
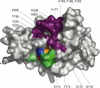
Ubiquitylation, the modification of cellular proteins by the covalent attachment of ubiquitin, is critical for diverse biological processes including cell cycle progression, signal transduction and stress response. This process can be reversed and regulated by a group of proteases called deubiquitylating enzymes (DUBs). Otubains are a recently identified family of DUBs that belong to the ovarian tumour (OTU) superfamily of proteins. Here, we report the first crystal structure of an OTU superfamily protein, otubain 2, at 2.1 A resolution and propose a model for otubain-ubiquitin binding on the basis of other DUB structures. Although otubain 2 is a member of the cysteine protease superfamily of folds, its crystal structure shows a novel fold for DUBs. Moreover, the active-site cleft is sterically occluded by a novel loop conformation resulting in an oxyanion hole, which consists uniquely of backbone amides, rather than the composite backbone/side-chain substructures seen in other DUBs and cysteine proteases. Furthermore, the residues that orient and stabilize the active-site histidine of otubain 2 are different from other cysteine proteases. This reorganization of the active-site topology provides a possible explanation for the low turnover and substrate specificity of the otubains.
Figures
Similar articles
-
Otubains: a new family of cysteine proteases in the ubiquitin pathway.EMBO Rep. 2003 May;4(5):517-22. doi: 10.1038/sj.embor.embor824. EMBO Rep. 2003. PMID: 12704427 Free PMC article.
-
Novel predicted peptidases with a potential role in the ubiquitin signaling pathway.Cell Cycle. 2004 Nov;3(11):1440-50. doi: 10.4161/cc.3.11.1206. Epub 2004 Nov 6. Cell Cycle. 2004. PMID: 15483401
-
Structural basis for Ufm1 processing by UfSP1.J Biol Chem. 2008 May 23;283(21):14893-900. doi: 10.1074/jbc.M708756200. Epub 2008 Mar 4. J Biol Chem. 2008. PMID: 18321862
-
The functions and regulation of Otubains in protein homeostasis and diseases.Ageing Res Rev. 2021 May;67:101303. doi: 10.1016/j.arr.2021.101303. Epub 2021 Feb 17. Ageing Res Rev. 2021. PMID: 33609777 Review.
-
Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes.Annu Rev Biochem. 2009;78:363-97. doi: 10.1146/annurev.biochem.78.082307.091526. Annu Rev Biochem. 2009. PMID: 19489724 Free PMC article. Review.
Cited by
-
OTU deubiquitinase, ubiquitin aldehyde binding 2 (OTUB2) modulates the stemness feature, chemoresistance, and epithelial-mesenchymal transition of colon cancer via regulating GINS complex subunit 1 (GINS1) expression.Cell Commun Signal. 2024 Aug 29;22(1):420. doi: 10.1186/s12964-024-01789-2. Cell Commun Signal. 2024. PMID: 39210373 Free PMC article.
-
Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein.J Biol Chem. 2008 Apr 18;283(16):11038-49. doi: 10.1074/jbc.M704398200. Epub 2008 Feb 12. J Biol Chem. 2008. PMID: 18270205 Free PMC article.
-
The porcine reproductive and respiratory syndrome virus nsp2 cysteine protease domain possesses both trans- and cis-cleavage activities.J Virol. 2009 Sep;83(18):9449-63. doi: 10.1128/JVI.00834-09. Epub 2009 Jul 8. J Virol. 2009. PMID: 19587037 Free PMC article.
-
Characterization of a novel otubain-like protease with deubiquitination activity from Nosema bombycis (Microsporidia).Parasitol Res. 2015 Oct;114(10):3759-66. doi: 10.1007/s00436-015-4624-7. Epub 2015 Jul 17. Parasitol Res. 2015. PMID: 26177898
-
Autophagy-Related Deubiquitinating Enzymes Involved in Health and Disease.Cells. 2015 Oct 5;4(4):596-621. doi: 10.3390/cells4040596. Cells. 2015. PMID: 26445063 Free PMC article. Review.
References
-
- Ben-Neriah Y (2002) Regulatory functions of ubiquitination in the immune system. Nat Immunol 3: 20–26 - PubMed
-
- Beyaert R, Heyninck K, Van Huffel S (2000) A20 and A20-binding proteins as cellular inhibitors of nuclear factor-κB-dependent gene expression and apoptosis. Biochem Pharmacol 60: 1143–1151 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

