Lineage-specific gene duplication and loss in human and great ape evolution
- PMID: 15252450
- PMCID: PMC449870
- DOI: 10.1371/journal.pbio.0020207
Lineage-specific gene duplication and loss in human and great ape evolution
Abstract
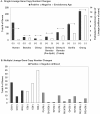
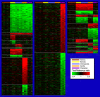
Given that gene duplication is a major driving force of evolutionary change and the key mechanism underlying the emergence of new genes and biological processes, this study sought to use a novel genome-wide approach to identify genes that have undergone lineage-specific duplications or contractions among several hominoid lineages. Interspecies cDNA array-based comparative genomic hybridization was used to individually compare copy number variation for 39,711 cDNAs, representing 29,619 human genes, across five hominoid species, including human. We identified 1,005 genes, either as isolated genes or in clusters positionally biased toward rearrangement-prone genomic regions, that produced relative hybridization signals unique to one or more of the hominoid lineages. Measured as a function of the evolutionary age of each lineage, genes showing copy number expansions were most pronounced in human (134) and include a number of genes thought to be involved in the structure and function of the brain. This work represents, to our knowledge, the first genome-wide gene-based survey of gene duplication across hominoid species. The genes identified here likely represent a significant majority of the major gene copy number changes that have occurred over the past 15 million years of human and great ape evolution and are likely to underlie some of the key phenotypic characteristics that distinguish these species.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures




Similar articles
-
Gene copy number variation spanning 60 million years of human and primate evolution.Genome Res. 2007 Sep;17(9):1266-77. doi: 10.1101/gr.6557307. Epub 2007 Jul 31. Genome Res. 2007. PMID: 17666543 Free PMC article.
-
Large-scale variation among human and great ape genomes determined by array comparative genomic hybridization.Genome Res. 2003 Mar;13(3):347-57. doi: 10.1101/gr.1003303. Genome Res. 2003. PMID: 12618365 Free PMC article.
-
Phylogenomic approaches to common problems encountered in the analysis of low copy repeats: the sulfotransferase 1A gene family example.BMC Evol Biol. 2005 Mar 7;5:22. doi: 10.1186/1471-2148-5-22. BMC Evol Biol. 2005. PMID: 15752422 Free PMC article.
-
Hox clusters as models for vertebrate genome evolution.Trends Genet. 2005 Aug;21(8):421-4. doi: 10.1016/j.tig.2005.06.004. Trends Genet. 2005. PMID: 15967537 Review.
-
The jewels of our genome: the search for the genomic changes underlying the evolutionarily unique capacities of the human brain.PLoS Genet. 2006 May;2(5):e80. doi: 10.1371/journal.pgen.0020080. PLoS Genet. 2006. PMID: 16733552 Free PMC article. Review.
Cited by
-
Evolution and diversity of copy number variation in the great ape lineage.Genome Res. 2013 Sep;23(9):1373-82. doi: 10.1101/gr.158543.113. Epub 2013 Jul 3. Genome Res. 2013. PMID: 23825009 Free PMC article.
-
The heterochromatic chromosome caps in great apes impact telomere metabolism.Nucleic Acids Res. 2013 May;41(9):4792-801. doi: 10.1093/nar/gkt169. Epub 2013 Mar 21. Nucleic Acids Res. 2013. PMID: 23519615 Free PMC article.
-
Online resources for genomic structural variation.Methods Mol Biol. 2012;838:273-89. doi: 10.1007/978-1-61779-507-7_13. Methods Mol Biol. 2012. PMID: 22228017 Free PMC article.
-
Estrogen receptor alpha gene amplification in breast cancer: 25 years of debate.World J Clin Oncol. 2016 Apr 10;7(2):160-73. doi: 10.5306/wjco.v7.i2.160. World J Clin Oncol. 2016. PMID: 27081639 Free PMC article. Review.
-
Noncoding sequences near duplicated genes evolve rapidly.Genome Biol Evol. 2010;2:518-33. doi: 10.1093/gbe/evq037. Epub 2010 Jun 29. Genome Biol Evol. 2010. PMID: 20660939 Free PMC article.
References
-
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, et al. Recent segmental duplications in the human genome. Science. 2002a;297:1003–1007. - PubMed
-
- Bieche I, Olivi M, Champeme MH, Vidaud D, Lidereau R, et al. Novel approach to quantitative polymerase chain reaction using real-time detection: Application to the detection of gene amplification in breast cancer. Int J Cancer. 1998;78:661–666. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

