Duplicated genes evolve slower than singletons despite the initial rate increase
- PMID: 15238160
- PMCID: PMC481058
- DOI: 10.1186/1471-2148-4-22
Duplicated genes evolve slower than singletons despite the initial rate increase
Abstract
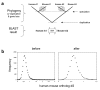
Background: Gene duplication is an important mechanism that can lead to the emergence of new functions during evolution. The impact of duplication on the mode of gene evolution has been the subject of several theoretical and empirical comparative-genomic studies. It has been shown that, shortly after the duplication, genes seem to experience a considerable relaxation of purifying selection.
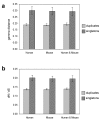
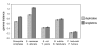
Results: Here we demonstrate two opposite effects of gene duplication on evolutionary rates. Sequence comparisons between paralogs show that, in accord with previous observations, a substantial acceleration in the evolution of paralogs occurs after duplication, presumably due to relaxation of purifying selection. The effect of gene duplication on evolutionary rate was also assessed by sequence comparison between orthologs that have paralogs (duplicates) and those that do not (singletons). It is shown that, in eukaryotes, duplicates, on average, evolve significantly slower than singletons. Eukaryotic ortholog evolutionary rates for duplicates are also negatively correlated with the number of paralogs per gene and the strength of selection between paralogs. A tally of annotated gene functions shows that duplicates tend to be enriched for proteins with known functions, particularly those involved in signaling and related cellular processes; by contrast, singletons include an over-abundance of poorly characterized proteins.
Conclusions: These results suggest that whether or not a gene duplicate is retained by selection depends critically on the pre-existing functional utility of the protein encoded by the ancestral singleton. Duplicates of genes of a higher biological import, which are subject to strong functional constraints on the sequence, are retained relatively more often. Thus, the evolutionary trajectory of duplicated genes appears to be determined by two opposing trends, namely, the post-duplication rate acceleration and the generally slow evolutionary rate owing to the high level of functional constraints.
Figures

Similar articles
-
Extensive divergence in alternative splicing patterns after gene and genome duplication during the evolutionary history of Arabidopsis.Mol Biol Evol. 2010 Jul;27(7):1686-97. doi: 10.1093/molbev/msq054. Epub 2010 Feb 25. Mol Biol Evol. 2010. PMID: 20185454
-
Not born equal: increased rate asymmetry in relocated and retrotransposed rodent gene duplicates.Mol Biol Evol. 2007 Mar;24(3):679-86. doi: 10.1093/molbev/msl199. Epub 2006 Dec 18. Mol Biol Evol. 2007. PMID: 17179139
-
The evolutionary fate of recently duplicated retrogenes in mice.J Evol Biol. 2007 Mar;20(2):617-26. doi: 10.1111/j.1420-9101.2006.01245.x. J Evol Biol. 2007. PMID: 17305828
-
Retention of protein complex membership by ancient duplicated gene products in budding yeast.Trends Genet. 2007 Jun;23(6):266-9. doi: 10.1016/j.tig.2007.03.012. Epub 2007 Apr 10. Trends Genet. 2007. PMID: 17428571 Review.
-
Diagnosing duplications--can it be done?Trends Genet. 2006 Mar;22(3):156-64. doi: 10.1016/j.tig.2006.01.002. Epub 2006 Jan 26. Trends Genet. 2006. PMID: 16442663 Review.
Cited by
-
Population transcriptomics uncovers the regulation of gene expression variation in adaptation to changing environment.Sci Rep. 2016 May 6;6:25536. doi: 10.1038/srep25536. Sci Rep. 2016. PMID: 27150248 Free PMC article.
-
Evolutionary dynamics of human autoimmune disease genes and malfunctioned immunological genes.BMC Evol Biol. 2012 Jan 25;12:10. doi: 10.1186/1471-2148-12-10. BMC Evol Biol. 2012. PMID: 22276655 Free PMC article.
-
Duplication and population dynamics shape historic patterns of selection and genetic variation at the major histocompatibility complex in rodents.Ecol Evol. 2013 Jun;3(6):1552-68. doi: 10.1002/ece3.567. Epub 2013 Apr 22. Ecol Evol. 2013. PMID: 23789067 Free PMC article.
-
Origins and impact of constraints in evolution of gene families.Genome Res. 2006 Dec;16(12):1529-36. doi: 10.1101/gr.5346206. Epub 2006 Oct 19. Genome Res. 2006. PMID: 17053091 Free PMC article.
-
A catalog of neutral and deleterious polymorphism in yeast.PLoS Genet. 2008 Aug 29;4(8):e1000183. doi: 10.1371/journal.pgen.1000183. PLoS Genet. 2008. PMID: 18769710 Free PMC article.
References
-
- Haldane JBS. The part played by recurrent mutation in evolution. Am Nat. 1933;67:5–19. doi: 10.1086/280465. - DOI
-
- Fisher RA. The possible modification of the response of the wild type to recurrent mutations. Am Nat. 1928;62:115–126. doi: 10.1086/280193. - DOI
-
- Pauling L, Zuckerkandl E. Chemical paleogenetics: molecular restoration studies of extinct forms of life. Acta Chem Scand. 1963;17:S9–S16.
-
- Ohno S. Evolution by gene duplication. Berlin-Heidelberg-New York, Springer-Verlag; 1970.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

