Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance
- PMID: 15215892
- PMCID: PMC449767
- DOI: 10.1038/sj.emboj.7600255
Human Orc2 localizes to centrosomes, centromeres and heterochromatin during chromosome inheritance
Erratum in
- EMBO J. 2005 Mar 9;24(5):1092-4
Abstract
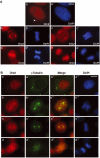
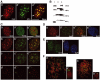
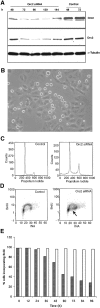
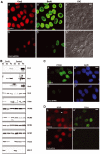
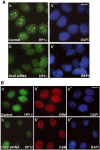
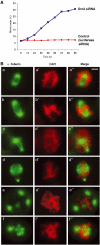
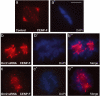
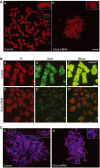
The initiation of DNA replication in S phase requires the prior assembly of an origin recognition complex (ORC)-dependent pre-replicative complex on chromatin during G1 phase of the cell division cycle. In human cells, the Orc2 subunit localized to the nucleus as expected, but it also localized to centrosomes throughout the entire cell cycle. Furthermore, Orc2 was tightly bound to heterochromatin and heterochromatin protein 1alpha (HP1alpha) and HP1beta in G1 and early S phase, but during late S, G2 and M phases tight chromatin association was restricted to centromeres. Depletion of Orc2 by siRNA caused multiple phenotypes. A population of cells showed an S-phase defect with little proliferating cell nuclear antigen (PCNA) on chromatin, although MCM proteins remained. Orc2 depletion also disrupted HP1 localization, but not histone-H3-lysine-9 methylation at prominent heterochromatic foci. Another subset of Orc2-depleted cells containing replicated DNA arrested with abnormally condensed chromosomes, failed chromosome congression and multiple centrosomes. These results implicate Orc2 protein in chromosome duplication, chromosome structure and centrosome copy number control, suggesting that it coordinates all stages of the chromosome inheritance cycle.
Figures
Similar articles
-
Human origin recognition complex is essential for HP1 binding to chromatin and heterochromatin organization.Proc Natl Acad Sci U S A. 2010 Aug 24;107(34):15093-8. doi: 10.1073/pnas.1009945107. Epub 2010 Aug 5. Proc Natl Acad Sci U S A. 2010. PMID: 20689044 Free PMC article.
-
Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase.J Cell Sci. 2003 Aug 15;116(Pt 16):3327-38. doi: 10.1242/jcs.00635. Epub 2003 Jul 2. J Cell Sci. 2003. PMID: 12840071
-
A CAF-1 dependent pool of HP1 during heterochromatin duplication.EMBO J. 2004 Sep 1;23(17):3516-26. doi: 10.1038/sj.emboj.7600362. Epub 2004 Aug 12. EMBO J. 2004. PMID: 15306854 Free PMC article.
-
Replication of heterochromatin: insights into mechanisms of epigenetic inheritance.Chromosoma. 2005 Dec;114(6):389-402. doi: 10.1007/s00412-005-0024-6. Epub 2005 Nov 15. Chromosoma. 2005. PMID: 16220346 Review.
-
Two distinct pathways responsible for the loading of CENP-A to centromeres in the fission yeast cell cycle.Philos Trans R Soc Lond B Biol Sci. 2005 Mar 29;360(1455):595-606; discussion 606-7. doi: 10.1098/rstb.2004.1614. Philos Trans R Soc Lond B Biol Sci. 2005. PMID: 15897182 Free PMC article. Review.
Cited by
-
Meier-Gorlin syndrome mutations disrupt an Orc1 CDK inhibitory domain and cause centrosome reduplication.Genes Dev. 2012 Aug 15;26(16):1797-810. doi: 10.1101/gad.197178.112. Epub 2012 Aug 1. Genes Dev. 2012. PMID: 22855792 Free PMC article.
-
New and Xisting regulatory mechanisms of X chromosome inactivation.Curr Opin Genet Dev. 2012 Apr;22(2):62-71. doi: 10.1016/j.gde.2012.02.007. Epub 2012 Mar 16. Curr Opin Genet Dev. 2012. PMID: 22424802 Free PMC article. Review.
-
A genome-wide map of DNA replication at single-molecule resolution in the malaria parasite Plasmodium falciparum.Nucleic Acids Res. 2023 Apr 11;51(6):2709-2724. doi: 10.1093/nar/gkad093. Nucleic Acids Res. 2023. PMID: 36808528 Free PMC article.
-
Regulation of DNA replication by chromatin structures: accessibility and recruitment.Chromosoma. 2011 Feb;120(1):39-46. doi: 10.1007/s00412-010-0287-4. Epub 2010 Aug 3. Chromosoma. 2011. PMID: 20680317 Review.
-
Nucleosome-interacting proteins regulated by DNA and histone methylation.Cell. 2010 Oct 29;143(3):470-84. doi: 10.1016/j.cell.2010.10.012. Cell. 2010. PMID: 21029866 Free PMC article.
References
-
- Allshire RC (1995) Elements of chromosome structure and function in fission yeast. Semin Cell Biol 6: 55–64 - PubMed
-
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 - PubMed
-
- Aparicio OM, Weinstein DM, Bell SP (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69 - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 - PubMed
-
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

