The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator
- PMID: 15215316
- PMCID: PMC515335
- DOI: 10.1091/mbc.e04-04-0293
The cochaperone HspBP1 inhibits the CHIP ubiquitin ligase and stimulates the maturation of the cystic fibrosis transmembrane conductance regulator
Abstract
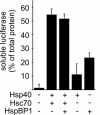
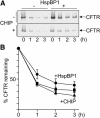
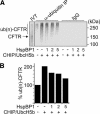
The CHIP ubiquitin ligase turns molecular chaperones into protein degradation factors. CHIP associates with the chaperones Hsc70 and Hsp90 during the regulation of signaling pathways and during protein quality control, and directs chaperone-bound clients to the proteasome for degradation. Obviously, this destructive activity should be carefully controlled. Here, we identify the cochaperone HspBP1 as an inhibitor of CHIP. HspBP1 attenuates the ubiquitin ligase activity of CHIP when complexed with Hsc70. As a consequence, HspBP1 interferes with the CHIP-induced degradation of immature forms of the cystic fibrosis transmembrane conductance regulator (CFTR) and stimulates CFTR maturation. Our data reveal a novel regulatory mechanism that determines folding and degradation activities of molecular chaperones.
Figures


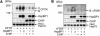



Similar articles
-
BAG-2 acts as an inhibitor of the chaperone-associated ubiquitin ligase CHIP.Mol Biol Cell. 2005 Dec;16(12):5891-900. doi: 10.1091/mbc.e05-07-0660. Epub 2005 Oct 5. Mol Biol Cell. 2005. PMID: 16207813 Free PMC article.
-
Human heat shock protein 105/110 kDa (Hsp105/110) regulates biogenesis and quality control of misfolded cystic fibrosis transmembrane conductance regulator at multiple levels.J Biol Chem. 2012 Jun 1;287(23):19158-70. doi: 10.1074/jbc.M111.297580. Epub 2012 Apr 13. J Biol Chem. 2012. PMID: 22505710 Free PMC article.
-
The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation.Nat Cell Biol. 2001 Jan;3(1):100-5. doi: 10.1038/35050509. Nat Cell Biol. 2001. PMID: 11146634
-
CHIP: a quality-control E3 ligase collaborating with molecular chaperones.Int J Biochem Cell Biol. 2003 May;35(5):572-8. doi: 10.1016/s1357-2725(02)00394-1. Int J Biochem Cell Biol. 2003. PMID: 12672450 Review.
-
Cystic fibrosis transmembrane conductance regulator degradation: cross-talk between the ubiquitylation and SUMOylation pathways.FEBS J. 2013 Sep;280(18):4430-8. doi: 10.1111/febs.12415. Epub 2013 Jul 22. FEBS J. 2013. PMID: 23809253 Free PMC article. Review.
Cited by
-
Stabilizing the Hsp70-Tau Complex Promotes Turnover in Models of Tauopathy.Cell Chem Biol. 2016 Aug 18;23(8):992-1001. doi: 10.1016/j.chembiol.2016.04.014. Epub 2016 Aug 4. Cell Chem Biol. 2016. PMID: 27499529 Free PMC article.
-
Molecular Chaperones as Targets to Circumvent the CFTR Defect in Cystic Fibrosis.Front Pharmacol. 2012 Jul 17;3:137. doi: 10.3389/fphar.2012.00137. eCollection 2012. Front Pharmacol. 2012. PMID: 22822398 Free PMC article.
-
Preventing illicit liaisons in Poland.EMBO Rep. 2005 Dec;6(12):1126-30. doi: 10.1038/sj.embor.7400581. EMBO Rep. 2005. PMID: 16299469 Free PMC article.
-
Small heat-shock proteins select deltaF508-CFTR for endoplasmic reticulum-associated degradation.Mol Biol Cell. 2007 Mar;18(3):806-14. doi: 10.1091/mbc.e06-05-0458. Epub 2006 Dec 20. Mol Biol Cell. 2007. PMID: 17182856 Free PMC article.
-
Hsp70 cochaperones HspBP1 and BAG-1M differentially regulate steroid hormone receptor function.PLoS One. 2014 Jan 14;9(1):e85415. doi: 10.1371/journal.pone.0085415. eCollection 2014. PLoS One. 2014. PMID: 24454860 Free PMC article.
References
-
- Alberti, S., Demand, J., Esser, C., Emmerich, N., Schild, H., and Höhfeld, J. (2002). Ubiquitylation of BAG-1 suggests a novel regulatory mechanism during the sorting of chaperone substrates to the proteasome. J. Biol. Chem. 277, 45920-45927. - PubMed
-
- Ballinger, C.A., Connell, P., Wu, Y., Hu, Z., Thompson, L.J., Yin, L.Y., and Patterson, C. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535-4545. - PMC - PubMed
-
- Cardozo, C.P., Michaud, C., Ost, M.C., Fliss, A.E., Yang, E., Patterson, C., Hall, S.J., and Caplan, A.J. (2003). C-terminal Hsp-interacting protein slows androgen receptor synthesis and reduces its rate of degradation. Arch. Biochem. Biophys. 410, 134-140. - PubMed
-
- Connell, P., Ballinger, C.A., Jiang, J., Wu, Y., Thompson, L.J., Höhfeld, J., and Patterson, C. (2001). The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat. Cell Biol. 3, 93-96. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

