Functional dissection of Escherichia coli trigger factor: unraveling the function of individual domains
- PMID: 15175291
- PMCID: PMC419933
- DOI: 10.1128/JB.186.12.3777-3784.2004
Functional dissection of Escherichia coli trigger factor: unraveling the function of individual domains
Abstract
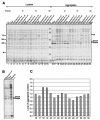
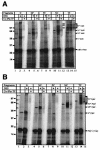
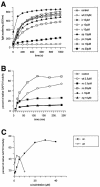
In Escherichia coli, the ribosome-associated chaperone Trigger Factor (TF) promotes the folding of newly synthesized cytosolic proteins. TF is composed of three domains: an N-terminal domain (N), which mediates ribosome binding; a central domain (P), which has peptidyl-prolyl cis/trans isomerase activity and is involved in substrate binding in vitro; and a C-terminal domain (C) with unknown function. We investigated the contributions of individual domains (N, P, and C) or domain combinations (NP, PC, and NC) to the chaperone activity of TF in vivo and in vitro. All fragments comprising the N domain (N, NP, NC) complemented the synthetic lethality of Deltatig DeltadnaK in cells lacking TF and DnaK, prevented protein aggregation in these cells, and cross-linked to nascent polypeptides in vitro. However, DeltatigDeltadnaK cells expressing the N domain alone grew more slowly and showed less viability than DeltatigDeltadnaK cells synthesizing either NP, NC, or full-length TF, indicating beneficial contributions of the P and C domains to TF's chaperone activity. In an in vitro system with purified components, none of the TF fragments assisted the refolding of denatured d-glyceraldehyde-3-phosphate dehydrogenase in a manner comparable to that of wild-type TF, suggesting that the observed chaperone activity of TF fragments in vivo is dependent on their localization at the ribosome. These results indicate that the N domain, in addition to its function to promote binding to the ribosome, has a chaperone activity per se and is sufficient to substitute for TF in vivo.
Figures


Similar articles
-
PPIase domain of trigger factor acts as auxiliary chaperone site to assist the folding of protein substrates bound to the crevice of trigger factor.Int J Biochem Cell Biol. 2010 Jun;42(6):890-901. doi: 10.1016/j.biocel.2010.01.019. Epub 2010 Jan 21. Int J Biochem Cell Biol. 2010. PMID: 20096367
-
Binding specificity of Escherichia coli trigger factor.Proc Natl Acad Sci U S A. 2001 Dec 4;98(25):14244-9. doi: 10.1073/pnas.261432298. Epub 2001 Nov 27. Proc Natl Acad Sci U S A. 2001. PMID: 11724963 Free PMC article.
-
Trigger Factor can antagonize both SecB and DnaK/DnaJ chaperone functions in Escherichia coli.Proc Natl Acad Sci U S A. 2007 Feb 27;104(9):3101-6. doi: 10.1073/pnas.0608232104. Epub 2007 Feb 20. Proc Natl Acad Sci U S A. 2007. PMID: 17360615 Free PMC article.
-
A cradle for new proteins: trigger factor at the ribosome.Curr Opin Struct Biol. 2005 Apr;15(2):204-12. doi: 10.1016/j.sbi.2005.03.005. Curr Opin Struct Biol. 2005. PMID: 15837180 Review.
-
Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding?Curr Opin Microbiol. 2003 Apr;6(2):157-62. doi: 10.1016/s1369-5274(03)00030-4. Curr Opin Microbiol. 2003. PMID: 12732306 Review.
Cited by
-
Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone.Cell. 2009 Sep 4;138(5):923-34. doi: 10.1016/j.cell.2009.07.044. Cell. 2009. PMID: 19737520 Free PMC article.
-
Molecular chaperones are nanomachines that catalytically unfold misfolded and alternatively folded proteins.Cell Mol Life Sci. 2014 Sep;71(17):3311-25. doi: 10.1007/s00018-014-1627-y. Epub 2014 Apr 24. Cell Mol Life Sci. 2014. PMID: 24760129 Free PMC article. Review.
-
Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo.Cell. 2011 Dec 9;147(6):1295-308. doi: 10.1016/j.cell.2011.10.044. Cell. 2011. PMID: 22153074 Free PMC article.
-
Regulation by a chaperone improves substrate selectivity during cotranslational protein targeting.Proc Natl Acad Sci U S A. 2015 Jun 23;112(25):E3169-78. doi: 10.1073/pnas.1422594112. Epub 2015 Jun 8. Proc Natl Acad Sci U S A. 2015. PMID: 26056263 Free PMC article.
-
Proteomic studies of an Antarctic cold-adapted bacterium, Shewanella livingstonensis Ac10, for global identification of cold-inducible proteins.Extremophiles. 2007 Nov;11(6):819-26. doi: 10.1007/s00792-007-0098-6. Epub 2007 Jul 7. Extremophiles. 2007. PMID: 17618403
References
-
- Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. - PubMed
-
- Callebaut, I., and J. P. Mornon. 1995. Trigger factor, one of the Escherichia coli chaperone proteins, is an original member of the FKBP family. FEBS Lett. 374:211-215. - PubMed
-
- Deuerling, E., H. Patzelt, S. Vorderwülbecke, T. Rauch, G. Kramer, E. Schaffitzel, A. Mogk, A. Schulze-Specking, H. Langen, and B. Bukau. 2003. Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol. Microbiol. 47:1317-1328. - PubMed
-
- Deuerling, E., A. Schulze-Specking, T. Tomoyasu, A. Mogk, and B. Bukau. 1999. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400:693-696. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

