The J-protein family: modulating protein assembly, disassembly and translocation
- PMID: 15170475
- PMCID: PMC1299080
- DOI: 10.1038/sj.embor.7400172
The J-protein family: modulating protein assembly, disassembly and translocation
Abstract
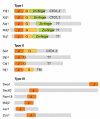
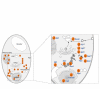
DnaJ is a molecular chaperone and the prototypical member of the J-protein family. J proteins are defined by the presence of a J domain that can regulate the activity of 70-kDa heat-shock proteins. Sequence analysis on the genome of Saccharomyces cerevisiae has revealed 22 proteins that establish four distinguishing structural features of the J domain: predicted helicity in segments I-IV, precisely placed interhelical contact residues, a lysine-rich surface on helix II and placement of the diagnostic sequence HPD between the predicted helices II and III. We suggest that this definition of the J-protein family could be used for other genome-wide studies. In addition, three J-like proteins were identified in yeast that contain regions closely resembling a J domain, but in which the HPD motif is non-conservatively replaced. We suggest that J-like proteins might function to regulate the activity of bona fide J proteins during protein translocation, assembly and disassembly.
Figures
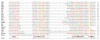
Similar articles
-
Rational mutagenesis of a 40 kDa heat shock protein from Agrobacterium tumefaciens identifies amino acid residues critical to its in vivo function.Int J Biochem Cell Biol. 2005 Jan;37(1):177-91. doi: 10.1016/j.biocel.2004.06.009. Int J Biochem Cell Biol. 2005. PMID: 15381160
-
NMR structure of the J-domain and the Gly/Phe-rich region of the Escherichia coli DnaJ chaperone.J Mol Biol. 1996 Jul 12;260(2):236-50. doi: 10.1006/jmbi.1996.0395. J Mol Biol. 1996. PMID: 8764403
-
Inactivation of the PKR protein kinase and stimulation of mRNA translation by the cellular co-chaperone P58(IPK) does not require J domain function.Biochemistry. 2002 Apr 16;41(15):4938-45. doi: 10.1021/bi0121499. Biochemistry. 2002. PMID: 11939789
-
Yeast prions help identify and define chaperone interaction networks.Curr Pharm Biotechnol. 2014;15(11):1008-18. doi: 10.2174/1389201015666141103021035. Curr Pharm Biotechnol. 2014. PMID: 25373385 Free PMC article. Review.
-
The molecular chaperone Hsp104--a molecular machine for protein disaggregation.J Struct Biol. 2006 Oct;156(1):139-48. doi: 10.1016/j.jsb.2006.02.004. Epub 2006 Mar 6. J Struct Biol. 2006. PMID: 16563798 Review.
Cited by
-
Eukaryotic DNAJ/K Database: A Comprehensive Phylogenomic Analysis Platform for the DNAJ/K Family.Genomics Inform. 2013 Mar;11(1):52-4. doi: 10.5808/GI.2013.11.1.52. Epub 2013 Mar 31. Genomics Inform. 2013. PMID: 23613683 Free PMC article.
-
Comparative proteomics in tall fescue to reveal underlying mechanisms for improving Photosystem II thermotolerance during heat stress memory.BMC Genomics. 2024 Jul 9;25(1):683. doi: 10.1186/s12864-024-10580-z. BMC Genomics. 2024. PMID: 38982385 Free PMC article.
-
Structure and mechanistic insights into novel iron-mediated moonlighting functions of human J-protein cochaperone, Dph4.J Biol Chem. 2012 Apr 13;287(16):13194-205. doi: 10.1074/jbc.M112.339655. Epub 2012 Feb 24. J Biol Chem. 2012. PMID: 22367199 Free PMC article.
-
Characterization of the human HSC20, an unusual DnaJ type III protein, involved in iron-sulfur cluster biogenesis.Hum Mol Genet. 2010 Oct 1;19(19):3816-34. doi: 10.1093/hmg/ddq301. Epub 2010 Jul 28. Hum Mol Genet. 2010. PMID: 20668094 Free PMC article.
-
The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex.Proc Natl Acad Sci U S A. 2005 Jul 19;102(29):10064-9. doi: 10.1073/pnas.0504400102. Epub 2005 Jul 7. Proc Natl Acad Sci U S A. 2005. PMID: 16002468 Free PMC article.
References
-
- Brodsky JL, Lawrence JG, Caplan AJ (1998) Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry 37: 18045–18055 - PubMed
-
- Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366 - PubMed
-
- Craig EA, Eisenman HC, Hundley HA (2003) Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding? Curr Opin Microbiol 6: 157–162 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

