The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation
- PMID: 15155881
- PMCID: PMC490034
- DOI: 10.1105/tpc.020958
The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation
Abstract
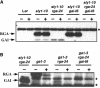
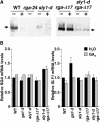
The nuclear DELLA proteins are highly conserved repressors of hormone gibberellin (GA) signaling in plants. In Arabidopsis thaliana, GA derepresses its signaling pathway by inducing proteolysis of the DELLA protein REPRESSOR OF ga1-3 (RGA). SLEEPY1 (SLY1) encodes an F-box-containing protein, and the loss-of-function sly1 mutant has a GA-insensitive dwarf phenotype and accumulates a high level of RGA. These findings suggested that SLY1 recruits RGA to the SCFSLY1 E3 ligase complex for ubiquitination and subsequent degradation by the 26S proteasome. In this report, we provide new insight into the molecular mechanism of how SLY1 interacts with the DELLA proteins for controlling GA response. By yeast two-hybrid and in vitro pull-down assays, we demonstrated that SLY1 interacts directly with RGA and GA INSENSITIVE (GAI, a closely related DELLA protein) via their C-terminal GRAS domain. The rga and gai null mutations additively suppressed the recessive sly1 mutant phenotype, further supporting the model that SCFSLY1 targets both RGA and GAI for degradation. The N-terminal DELLA domain of RGA previously was shown to be essential for GA-induced degradation. However, we found that this DELLA domain is not required for protein-protein interaction with SLY1 in yeast (Saccharomyces cerevisiae), suggesting that its role is in a GA-triggered conformational change of the DELLA proteins. We also identified a novel gain-of-function sly1-d mutation that increased GA signaling by reducing the levels of the DELLA protein in plants. This effect of sly1-d appears to be caused by an enhanced interaction between sly1-d and the DELLA proteins.
Copyright 2004 American Society of Plant Biologists
Figures




Similar articles
-
Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY.Proc Natl Acad Sci U S A. 2004 Aug 24;101(34):12771-6. doi: 10.1073/pnas.0404287101. Epub 2004 Aug 12. Proc Natl Acad Sci U S A. 2004. PMID: 15308775 Free PMC article.
-
The role of two f-box proteins, SLEEPY1 and SNEEZY, in Arabidopsis gibberellin signaling.Plant Physiol. 2011 Feb;155(2):765-75. doi: 10.1104/pp.110.166272. Epub 2010 Dec 16. Plant Physiol. 2011. PMID: 21163960 Free PMC article.
-
The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase.Plant Cell. 2003 May;15(5):1120-30. doi: 10.1105/tpc.010827. Plant Cell. 2003. PMID: 12724538 Free PMC article.
-
Molecular biology of gibberellins signaling in higher plants.Int Rev Cell Mol Biol. 2008;268:191-221. doi: 10.1016/S1937-6448(08)00806-X. Int Rev Cell Mol Biol. 2008. PMID: 18703407 Review.
-
Molecular mechanism of gibberellin signaling in plants.Annu Rev Plant Biol. 2004;55:197-223. doi: 10.1146/annurev.arplant.55.031903.141753. Annu Rev Plant Biol. 2004. PMID: 15377219 Review.
Cited by
-
Gibberellins and abscisic acid signal crosstalk: living and developing under unfavorable conditions.Plant Cell Rep. 2013 Jul;32(7):1007-16. doi: 10.1007/s00299-013-1409-2. Epub 2013 Mar 23. Plant Cell Rep. 2013. PMID: 23525744 Review.
-
Expression of the DELLA repressor GAI and its regulators SPY and SEC are impacted by disruption of chromatin modifiers.MicroPubl Biol. 2019 Oct 17;2019:10.17912/micropub.biology.000175. doi: 10.17912/micropub.biology.000175. MicroPubl Biol. 2019. PMID: 32550419 Free PMC article. No abstract available.
-
Mathematical modeling elucidates the role of transcriptional feedback in gibberellin signaling.Proc Natl Acad Sci U S A. 2012 May 8;109(19):7571-6. doi: 10.1073/pnas.1113666109. Epub 2012 Apr 20. Proc Natl Acad Sci U S A. 2012. PMID: 22523240 Free PMC article.
-
Beyond the Genetic Pathways, Flowering Regulation Complexity in Arabidopsis thaliana.Int J Mol Sci. 2021 May 27;22(11):5716. doi: 10.3390/ijms22115716. Int J Mol Sci. 2021. PMID: 34071961 Free PMC article. Review.
-
'Overgrowth' mutants in barley and wheat: new alleles and phenotypes of the 'Green Revolution' DELLA gene.J Exp Bot. 2013 Apr;64(6):1603-13. doi: 10.1093/jxb/ert022. Epub 2013 Feb 4. J Exp Bot. 2013. PMID: 23382550 Free PMC article.
References
-
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. - PubMed
-
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., and Smith, J.A. and Struhl, K., eds (1990). Current Protocols in Molecular Biology. (New York: Green Publishing Associates/Wiley-Interscience).
-
- Boss, P.K., and Thomas, M.R. (2002). Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416, 847–850. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

