Predicting transmembrane beta-barrels in proteomes
- PMID: 15141026
- PMCID: PMC419468
- DOI: 10.1093/nar/gkh580
Predicting transmembrane beta-barrels in proteomes
Abstract
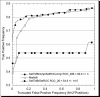
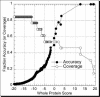
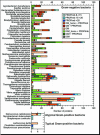
Very few methods address the problem of predicting beta-barrel membrane proteins directly from sequence. One reason is that only very few high-resolution structures for transmembrane beta-barrel (TMB) proteins have been determined thus far. Here we introduced the design, statistics and results of a novel profile-based hidden Markov model for the prediction and discrimination of TMBs. The method carefully attempts to avoid over-fitting the sparse experimental data. While our model training and scoring procedures were very similar to a recently published work, the architecture and structure-based labelling were significantly different. In particular, we introduced a new definition of beta- hairpin motifs, explicit state modelling of transmembrane strands, and a log-odds whole-protein discrimination score. The resulting method reached an overall four-state (up-, down-strand, periplasmic-, outer-loop) accuracy as high as 86%. Furthermore, accurately discriminated TMB from non-TMB proteins (45% coverage at 100% accuracy). This high precision enabled the application to 72 entirely sequenced Gram-negative bacteria. We found over 164 previously uncharacterized TMB proteins at high confidence. Database searches did not implicate any of these proteins with membranes. We challenge that the vast majority of our 164 predictions will eventually be verified experimentally. All proteome predictions and the PROFtmb prediction method are available at http://www.rostlab.org/ services/PROFtmb/.
Figures

Similar articles
-
PROFtmb: a web server for predicting bacterial transmembrane beta barrel proteins.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W186-8. doi: 10.1093/nar/gkl262. Nucleic Acids Res. 2006. PMID: 16844988 Free PMC article.
-
TMB-Hunt: an amino acid composition based method to screen proteomes for beta-barrel transmembrane proteins.BMC Bioinformatics. 2005 Mar 15;6:56. doi: 10.1186/1471-2105-6-56. BMC Bioinformatics. 2005. PMID: 15769290 Free PMC article.
-
Evaluation of methods for predicting the topology of beta-barrel outer membrane proteins and a consensus prediction method.BMC Bioinformatics. 2005 Jan 12;6:7. doi: 10.1186/1471-2105-6-7. BMC Bioinformatics. 2005. PMID: 15647112 Free PMC article.
-
Combined prediction of transmembrane topology and signal peptide of beta-barrel proteins: using a hidden Markov model and genetic algorithms.Comput Biol Med. 2010 Jul;40(7):621-8. doi: 10.1016/j.compbiomed.2010.04.006. Epub 2010 May 21. Comput Biol Med. 2010. PMID: 20488436
-
A sequence-profile-based HMM for predicting and discriminating beta barrel membrane proteins.Bioinformatics. 2002;18 Suppl 1:S46-53. doi: 10.1093/bioinformatics/18.suppl_1.s46. Bioinformatics. 2002. PMID: 12169530
Cited by
-
Trimeric structure of major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis.J Bacteriol. 2005 Feb;187(3):902-11. doi: 10.1128/JB.187.3.902-911.2005. J Bacteriol. 2005. PMID: 15659668 Free PMC article.
-
Evidence for the recent origin of a bacterial protein-coding, overlapping orphan gene by evolutionary overprinting.BMC Evol Biol. 2015 Dec 18;15:283. doi: 10.1186/s12862-015-0558-z. BMC Evol Biol. 2015. PMID: 26677845 Free PMC article.
-
Computational prediction of secreted proteins in gram-negative bacteria.Comput Struct Biotechnol J. 2021 Mar 22;19:1806-1828. doi: 10.1016/j.csbj.2021.03.019. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 33897982 Free PMC article. Review.
-
Three-Dimensional Structure Characterization and Inhibition Study of Exfoliative Toxin D From Staphylococcus aureus.Front Pharmacol. 2022 Feb 18;13:800970. doi: 10.3389/fphar.2022.800970. eCollection 2022. Front Pharmacol. 2022. PMID: 35250557 Free PMC article.
-
An automatic method for identifying surface proteins in bacteria: SLEP.BMC Bioinformatics. 2010 Jan 20;11:39. doi: 10.1186/1471-2105-11-39. BMC Bioinformatics. 2010. PMID: 20089159 Free PMC article.
References
-
- Schulz G.E. (2000) beta-Barrel membrane proteins. Curr. Opin. Struct. Biol., 10, 443–447. - PubMed
-
- Pautsch A. and Schulz,G.E. (2000) High-resolution structure of the OmpA membrane domain. J. Mol. Biol., 298, 273–282. - PubMed
-
- Forst D., Welte,W., Wacker,T. and Diederichs,K. (1998) Structure of the sucrose-specific porin ScrY from Salmonella typhimurium and its complex with sucrose. Nature Struct. Biol., 5, 37–46. - PubMed
-
- Wang Y.F., Dutzler,R., Rizkallah,P.J., Rosenbusch,J.P. and Schirmer,T. (1997) Channel specificity: structural basis for sugar discrimination and differential flux rates in maltoporin. J. Mol. Biol., 272, 56–63. - PubMed
-
- Koebnik R., Locher,K.P. and Van Gelder,P. (2000) Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Mol. Microbiol., 37, 239–253. - PubMed

