Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion
- PMID: 15140992
- PMCID: PMC415818
- DOI: 10.1128/JVI.78.11.5946-5956.2004
Mutational analyses of Epstein-Barr virus glycoprotein 42 reveal functional domains not involved in receptor binding but required for membrane fusion
Abstract
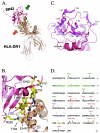
Epstein-Barr virus (EBV) is a human gammaherpesvirus associated with malignancies of both epithelial and lymphoid origin. Efficient infection of the latent host reservoir B lymphocytes involves the binding of glycoproteins gp350/220 for initial attachment, followed by the concerted action of gH, gL, gB, and gp42 for membrane fusion. The type II membrane protein gp42 is required for infection of B cells and assembles into a complex with gH and gL. The cellular host receptor for gp42, class II human leukocyte antigen (HLA), has been structurally verified by crystallization analyses of gp42 bound to HLA-DR1. Interestingly, the crystal structure revealed a hydrophobic pocket consisting of many aromatic and aliphatic residues from the predicted C-type lectin domain of gp42 that in other members of the C-type lectin family binds major histocompatibility complex class I or other diverse ligands. Although the hydrophobic pocket does not bind HLA class II, mutational analyses presented here indicate that this domain is essential for EBV-induced membrane fusion. In addition, mutational analysis of the region of gp42 contacting HLA class II in the gp42-HLA-DR1 cocrystal confirms that this region interacts with HLA class II and that this interaction is also important for EBV-induced membrane fusion.
Figures


Similar articles
-
Cleavage and secretion of Epstein-Barr virus glycoprotein 42 promote membrane fusion with B lymphocytes.J Virol. 2009 Jul;83(13):6664-72. doi: 10.1128/JVI.00195-09. Epub 2009 Apr 15. J Virol. 2009. PMID: 19369343 Free PMC article.
-
Membrane anchoring of Epstein-Barr virus gp42 inhibits fusion with B cells even with increased flexibility allowed by engineered spacers.mBio. 2015 Jan 6;6(1):e02285-14. doi: 10.1128/mBio.02285-14. mBio. 2015. PMID: 25564465 Free PMC article.
-
Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1.Mol Cell. 2002 Feb;9(2):375-85. doi: 10.1016/s1097-2765(02)00465-3. Mol Cell. 2002. PMID: 11864610
-
Structural and Mechanistic Insights into the Tropism of Epstein-Barr Virus.Mol Cells. 2016 Apr 30;39(4):286-91. doi: 10.14348/molcells.2016.0066. Epub 2016 Apr 6. Mol Cells. 2016. PMID: 27094060 Free PMC article. Review.
-
Integrins as triggers of Epstein-Barr virus fusion and epithelial cell infection.Virulence. 2010 Sep-Oct;1(5):395-8. doi: 10.4161/viru.1.5.12546. Virulence. 2010. PMID: 21178476 Free PMC article. Review.
Cited by
-
Cleavage of Epstein-Barr virus glycoprotein B is required for full function in cell-cell fusion with both epithelial and B cells.J Gen Virol. 2009 Mar;90(Pt 3):591-595. doi: 10.1099/vir.0.007237-0. J Gen Virol. 2009. PMID: 19218203 Free PMC article.
-
Neutralizing antibodies against EBV gp42 show potent in vivo protection and define novel epitopes.Emerg Microbes Infect. 2023 Dec;12(2):2245920. doi: 10.1080/22221751.2023.2245920. Emerg Microbes Infect. 2023. PMID: 37542379 Free PMC article.
-
Binding-site interactions between Epstein-Barr virus fusion proteins gp42 and gH/gL reveal a peptide that inhibits both epithelial and B-cell membrane fusion.J Virol. 2007 Sep;81(17):9216-29. doi: 10.1128/JVI.00575-07. Epub 2007 Jun 20. J Virol. 2007. PMID: 17581996 Free PMC article.
-
Fusion of epithelial cells by Epstein-Barr virus proteins is triggered by binding of viral glycoproteins gHgL to integrins alphavbeta6 or alphavbeta8.Proc Natl Acad Sci U S A. 2009 Dec 1;106(48):20464-9. doi: 10.1073/pnas.0907508106. Epub 2009 Nov 17. Proc Natl Acad Sci U S A. 2009. PMID: 19920174 Free PMC article.
-
Epstein-Barr virus gp42 is posttranslationally modified to produce soluble gp42 that mediates HLA class II immune evasion.J Virol. 2005 Jan;79(2):841-52. doi: 10.1128/JVI.79.2.841-852.2005. J Virol. 2005. PMID: 15613312 Free PMC article.
References
-
- Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, et al. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207-211. - PubMed
-
- Borza, C. M., and L. M. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. - PubMed
-
- Burkitt, D. 1962. A children's cancer dependent on climatic factors. Nauchni. Tr. Vissh. Med. Inst. Sofiia 194:232-234. - PubMed
-
- Carfí, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM61050/GM/NIGMS NIH HHS/United States
- R01 CA093444/CA/NCI NIH HHS/United States
- R01 AI038972/AI/NIAID NIH HHS/United States
- R01 GM061050/GM/NIGMS NIH HHS/United States
- T32 GM08061/GM/NIGMS NIH HHS/United States
- R56 AI038972/AI/NIAID NIH HHS/United States
- T32 GM008061/GM/NIGMS NIH HHS/United States
- R01 CA062234/CA/NCI NIH HHS/United States
- R01 DE013127/DE/NIDCR NIH HHS/United States
- CA62234/CA/NCI NIH HHS/United States
- CA93444/CA/NCI NIH HHS/United States
- R01 CA073507/CA/NCI NIH HHS/United States
- CA73507/CA/NCI NIH HHS/United States
- AI38972/AI/NIAID NIH HHS/United States
- DE13127/DE/NIDCR NIH HHS/United States
- R37 AI038972/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

