Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase
- PMID: 15140959
- PMCID: PMC415832
- DOI: 10.1128/JVI.78.11.5619-5632.2004
Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase
Abstract
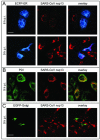
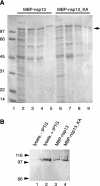
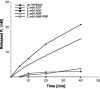
Severe acute respiratory syndrome coronavirus (SARS-CoV), a newly identified group 2 coronavirus, is the causative agent of severe acute respiratory syndrome, a life-threatening form of pneumonia in humans. Coronavirus replication and transcription are highly specialized processes of cytoplasmic RNA synthesis that localize to virus-induced membrane structures and were recently proposed to involve a complex enzymatic machinery that, besides RNA-dependent RNA polymerase, helicase, and protease activities, also involves a series of RNA-processing enzymes that are not found in most other RNA virus families. Here, we characterized the enzymatic activities of a recombinant form of the SARS-CoV helicase (nonstructural protein [nsp] 13), a superfamily 1 helicase with an N-terminal zinc-binding domain. We report that nsp13 has both RNA and DNA duplex-unwinding activities. SARS-CoV nsp13 unwinds its substrates in a 5'-to-3' direction and features a remarkable processivity, allowing efficient strand separation of extended regions of double-stranded RNA and DNA. Characterization of the nsp13-associated (deoxy)nucleoside triphosphatase ([dNTPase) activities revealed that all natural nucleotides and deoxynucleotides are substrates of nsp13, with ATP, dATP, and GTP being hydrolyzed slightly more efficiently than other nucleotides. Furthermore, we established an RNA 5'-triphosphatase activity for the SARS-CoV nsp13 helicase which may be involved in the formation of the 5' cap structure of viral RNAs. The data suggest that the (d)NTPase and RNA 5'-triphosphatase activities of nsp13 have a common active site. Finally, we established that, in SARS-CoV-infected Vero E6 cells, nsp13 localizes to membranes that appear to be derived from the endoplasmic reticulum and are the likely site of SARS-CoV RNA synthesis.
Figures





Similar articles
-
Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5'-triphosphatase activities.J Virol. 2004 Jul;78(14):7833-8. doi: 10.1128/JVI.78.14.7833-7838.2004. J Virol. 2004. PMID: 15220459 Free PMC article.
-
A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase nsP13 in the unwinding of duplex RNA.Sci Rep. 2020 Mar 11;10(1):4481. doi: 10.1038/s41598-020-61432-1. Sci Rep. 2020. PMID: 32161317 Free PMC article.
-
The severe acute respiratory syndrome (SARS) coronavirus NTPase/helicase belongs to a distinct class of 5' to 3' viral helicases.J Biol Chem. 2003 Oct 10;278(41):39578-82. doi: 10.1074/jbc.C300328200. Epub 2003 Aug 13. J Biol Chem. 2003. PMID: 12917423 Free PMC article.
-
The Coronavirus helicase in replication.Virus Res. 2024 Aug;346:199401. doi: 10.1016/j.virusres.2024.199401. Epub 2024 May 31. Virus Res. 2024. PMID: 38796132 Free PMC article. Review.
-
SARS-CoV ORF1b-encoded nonstructural proteins 12-16: replicative enzymes as antiviral targets.Antiviral Res. 2014 Jan;101:122-30. doi: 10.1016/j.antiviral.2013.11.006. Epub 2013 Nov 20. Antiviral Res. 2014. PMID: 24269475 Free PMC article. Review.
Cited by
-
Variable Inhibition of DNA Unwinding Rates Catalyzed by the SARS-CoV-2 Helicase Nsp13 by Structurally Distinct Single DNA Lesions.Int J Mol Sci. 2024 Jul 19;25(14):7930. doi: 10.3390/ijms25147930. Int J Mol Sci. 2024. PMID: 39063172 Free PMC article.
-
Inflammation in the COVID-19 airway is due to inhibition of CFTR signaling by the SARS-CoV-2 spike protein.Sci Rep. 2024 Jul 23;14(1):16895. doi: 10.1038/s41598-024-66473-4. Sci Rep. 2024. PMID: 39043712 Free PMC article.
-
Evidence of mitochondria origin of SARS-CoV-2 double-membrane vesicles: a review.F1000Res. 2024 Jul 10;10:1009. doi: 10.12688/f1000research.73170.2. eCollection 2021. F1000Res. 2024. PMID: 38827572 Free PMC article. Review.
-
Natural compounds may contribute in preventing SARS-CoV-2 infection: a narrative review.Food Sci Hum Wellness. 2022 Sep;11(5):1134-1142. doi: 10.1016/j.fshw.2022.04.005. Epub 2022 Jun 2. Food Sci Hum Wellness. 2022. PMID: 38621001 Free PMC article. Review.
-
Drug repurposing screen to identify inhibitors of the RNA polymerase (nsp12) and helicase (nsp13) from SARS-CoV-2 replication and transcription complex.Virus Res. 2024 May;343:199356. doi: 10.1016/j.virusres.2024.199356. Epub 2024 Mar 16. Virus Res. 2024. PMID: 38490582 Free PMC article.
References
-
- Anand, K., J. Ziebuhr, P. Wadhwani, J. R. Mesters, and R. Hilgenfeld. 2003. Coronavirus main proteinase (3CLpro) structure: basis for design of anti-SARS drugs. Science 300:1763-1767. - PubMed
-
- Bird, L. E., H. S. Subramanya, and D. B. Wigley. 1998. Helicases: a unifying structural theme? Curr. Opin. Struct. Biol. 8:14-18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

