Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome
- PMID: 15124102
- PMCID: PMC1182084
- DOI: 10.1086/421532
Mutations of the ephrin-B1 gene cause craniofrontonasal syndrome
Abstract
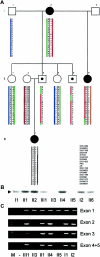
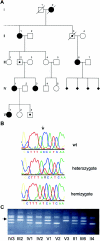
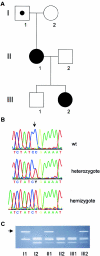
Craniofrontonasal syndrome (CFNS) is an X-linked craniofacial disorder with an unusual manifestation pattern, in which affected females show multiple skeletal malformations, whereas the genetic defect causes no or only mild abnormalities in male carriers. Recently, we have mapped a gene for CFNS in the pericentromeric region of the X chromosome that contains the EFNB1 gene, which encodes the ephrin-B1 ligand for Eph receptors. Since Efnb1 mutant mice display a spectrum of malformations and an unusual inheritance reminiscent of CFNS, we analyzed the EFNB1 gene in three families with CFNS. In one family, a deletion of exons 2-5 was identified in an obligate carrier male, his mildly affected brother, and in the affected females. In the two other families, missense mutations in EFNB1 were detected that lead to amino acid exchanges P54L and T111I. Both mutations are located in multimerization and receptor-interaction motifs found within the ephrin-B1 extracellular domain. In all cases, mutations were found consistently in obligate male carriers, clinically affected males, and affected heterozygous females. We conclude that mutations in EFNB1 cause CFNS.
Figures


Similar articles
-
The impact of CFNS-causing EFNB1 mutations on ephrin-B1 function.BMC Med Genet. 2010 Jun 17;11:98. doi: 10.1186/1471-2350-11-98. BMC Med Genet. 2010. PMID: 20565770 Free PMC article.
-
Twenty-six novel EFNB1 mutations in familial and sporadic craniofrontonasal syndrome (CFNS).Hum Mutat. 2005 Aug;26(2):113-8. doi: 10.1002/humu.20193. Hum Mutat. 2005. PMID: 15959873
-
Mutations of ephrin-B1 (EFNB1), a marker of tissue boundary formation, cause craniofrontonasal syndrome.Proc Natl Acad Sci U S A. 2004 Jun 8;101(23):8652-7. doi: 10.1073/pnas.0402819101. Epub 2004 May 27. Proc Natl Acad Sci U S A. 2004. PMID: 15166289 Free PMC article.
-
Clinical and genetic aspects of craniofrontonasal syndrome: towards resolving a genetic paradox.Mol Genet Metab. 2005 Sep-Oct;86(1-2):110-6. doi: 10.1016/j.ymgme.2005.07.017. Mol Genet Metab. 2005. PMID: 16143553 Review.
-
Diverse clinical and genetic aspects of craniofrontonasal syndrome.Pediatr Neurol. 2011 Feb;44(2):83-7. doi: 10.1016/j.pediatrneurol.2010.10.012. Pediatr Neurol. 2011. PMID: 21215906 Review.
Cited by
-
Presenilin-dependent intramembrane cleavage of ephrin-B1.Mol Neurodegener. 2006 Jun 12;1:2. doi: 10.1186/1750-1326-1-2. Mol Neurodegener. 2006. PMID: 16930449 Free PMC article.
-
Early onset X-linked female limited high myopia in three multigenerational families caused by novel mutations in the ARR3 gene.Hum Mutat. 2022 Mar;43(3):380-388. doi: 10.1002/humu.24327. Epub 2022 Jan 19. Hum Mutat. 2022. PMID: 35001458 Free PMC article.
-
Eph receptors and ephrin signaling pathways: a role in bone homeostasis.Int J Med Sci. 2008 Sep 3;5(5):263-72. doi: 10.7150/ijms.5.263. Int J Med Sci. 2008. PMID: 18797510 Free PMC article. Review.
-
Genetic Causes of Craniosynostosis: An Update.Mol Syndromol. 2019 Feb;10(1-2):6-23. doi: 10.1159/000492266. Epub 2018 Aug 15. Mol Syndromol. 2019. PMID: 30976276 Free PMC article. Review.
-
Crucial and Overlapping Roles of Six1 and Six2 in Craniofacial Development.J Dent Res. 2019 May;98(5):572-579. doi: 10.1177/0022034519835204. Epub 2019 Mar 24. J Dent Res. 2019. PMID: 30905259 Free PMC article.
References
Electronic-Database Information
-
- Ensembl Genome Server, http://www.ensembl.org (for EFNB1 [accession number ENSP00000204961])
-
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank (for EFNB1 reference sequence [accession number XM_038809])
-
- LocusLink, http://www.ncbi.nlm.nih.gov/LocusLink (for EFNB1 [accession number 1947])
-
- NCBI Molecular Variation Database (dbSNP), http://www.ncbi.nlm.nih.gov/SNP
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim
References
-
- Devriendt K, van Mol C, Fryns JP (1995) Craniofacial dysplasia: a more severe expression in the mother than in her son. Genet Couns 6:361–364 - PubMed
-
- Feldman GJ, Ward DE, Lajeunie-Renier E, Saavedra D, Robin NH, Proud V, Robb LJ, Der Kaloustian V, Carey JC, Cohen M, Cornier V, Munnich A, Zackai EH, Wilkie AOM, Price RA, Muenke M (1997) A novel phenotypic pattern in X-linked inheritance: craniofrontonasal syndrome maps to Xp22. Hum Mol Genet 6:1937–194110.1093/hmg/6.11.1937 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

