Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+
- PMID: 15122879
- PMCID: PMC2426922
- DOI: 10.1021/bi035213q
Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mn2+
Abstract
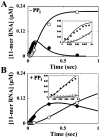
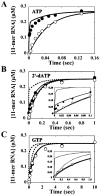
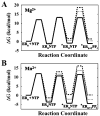
The use of Mn(2+) as the divalent cation cofactor in polymerase-catalyzed reactions instead of Mg(2+) often diminishes the stringency of substrate selection and incorporation fidelity. We have solved the complete kinetic mechanism for single nucleotide incorporation catalyzed by the RNA-dependent RNA polymerase from poliovirus (3D(pol)) in the presence of Mn(2+). The steps employed during a single cycle of nucleotide incorporation are identical to those employed in the presence of Mg(2+) and include a conformational-change step after nucleotide binding to achieve catalytic competence of the polymerase-primer/template-nucleotide complex. In the presence of Mn(2+), the conformational-change step is the primary determinant of enzyme specificity, phosphoryl transfer appears as the sole rate-limiting step for nucleotide incorporation, and the rate of phosphoryl transfer is the same for all nucleotides: correct and incorrect. Because phosphoryl transfer is the rate-limiting step in the presence of Mn(2+), it was possible to determine that the maximal phosphorothioate effect in this system is in the range of 8-11. This information permitted further interrogation of the nucleotide-selection process in the presence of Mg(2+), highlighting the capacity of this cation to permit the enzyme to use the phosphoryl-transfer step for nucleotide selection. The inability of Mn(2+) to support a reduction in the efficiency of phosphoryl transfer when incorrect substrates are employed is the primary explanation for the loss of fidelity observed in the presence of this cofactor. We propose that the conformational change involves reorientation of the triphosphate moiety of the bound nucleotide into a conformation that permits binding of the second metal ion required for catalysis. In the presence of Mg(2+), this conformation requires interactions with the enzyme that permit a reduction in catalytic efficiency to occur during an attempt to incorporate an incorrect nucleotide. Adventitious interactions in the cofactor-binding site with bound Mn(2+) may diminish fidelity by compensating for interaction losses used to modulate catalytic efficiency when incorrect nucleotides are bound in the presence of Mg(2+).
Figures

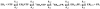
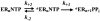
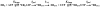
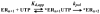
Similar articles
-
Poliovirus RNA-dependent RNA polymerase (3Dpol): pre-steady-state kinetic analysis of ribonucleotide incorporation in the presence of Mg2+.Biochemistry. 2004 May 11;43(18):5126-37. doi: 10.1021/bi035212y. Biochemistry. 2004. PMID: 15122878 Free PMC article.
-
Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection.Biochemistry. 2004 May 11;43(18):5149-58. doi: 10.1021/bi035429s. Biochemistry. 2004. PMID: 15122880 Free PMC article.
-
Poliovirus RNA-dependent RNA polymerase (3D(pol)). Divalent cation modulation of primer, template, and nucleotide selection.J Biol Chem. 1999 Dec 24;274(52):37060-9. doi: 10.1074/jbc.274.52.37060. J Biol Chem. 1999. PMID: 10601264
-
Fidelity of Nucleotide Incorporation by the RNA-Dependent RNA Polymerase from Poliovirus.Enzymes. 2016;39:293-323. doi: 10.1016/bs.enz.2016.02.002. Epub 2016 Mar 28. Enzymes. 2016. PMID: 27241934 Free PMC article. Review.
-
Incorporation fidelity of the viral RNA-dependent RNA polymerase: a kinetic, thermodynamic and structural perspective.Virus Res. 2005 Feb;107(2):141-9. doi: 10.1016/j.virusres.2004.11.004. Virus Res. 2005. PMID: 15649560 Free PMC article. Review.
Cited by
-
What is the role of motif D in the nucleotide incorporation catalyzed by the RNA-dependent RNA polymerase from poliovirus?PLoS Comput Biol. 2012;8(12):e1002851. doi: 10.1371/journal.pcbi.1002851. Epub 2012 Dec 27. PLoS Comput Biol. 2012. PMID: 23300428 Free PMC article.
-
Emergency Services of Viral RNAs: Repair and Remodeling.Microbiol Mol Biol Rev. 2018 Mar 14;82(2):e00067-17. doi: 10.1128/MMBR.00067-17. Print 2018 Jun. Microbiol Mol Biol Rev. 2018. PMID: 29540453 Free PMC article. Review.
-
Residues of the rotavirus RNA-dependent RNA polymerase template entry tunnel that mediate RNA recognition and genome replication.J Virol. 2011 Mar;85(5):1958-69. doi: 10.1128/JVI.01689-10. Epub 2010 Dec 8. J Virol. 2011. PMID: 21147920 Free PMC article.
-
Interstitial contacts in an RNA-dependent RNA polymerase lattice.J Mol Biol. 2011 Sep 30;412(4):737-50. doi: 10.1016/j.jmb.2011.07.053. Epub 2011 Aug 3. J Mol Biol. 2011. PMID: 21839092 Free PMC article.
-
Mechanisms of activity and inhibition of the hepatitis C virus RNA-dependent RNA polymerase.J Biol Chem. 2010 Apr 30;285(18):13685-93. doi: 10.1074/jbc.M109.082206. Epub 2010 Mar 1. J Biol Chem. 2010. PMID: 20194503 Free PMC article.
References
-
- Brautigam CA, Steitz TA. Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 1998;8:54–63. - PubMed
-
- Kornberg A, Baker T. DNA Replication. 2nd ed. W. H. Freeman and Co.; New York: 1991.
-
- Grabbara S, Peliska JA. Catalytic activities associated with retroviral and viral polymerases. Methods Enzymol. 1996;275:276–310. - PubMed
-
- Huang Y, Beaudry A, McSwiggen J, Sousa R. Determinants of ribose specificity in RNA polymerization: effects of Mn2+ and deoxynucleoside monophosphate incorporation into transcripts. Biochemistry. 1997;36:13718–13728. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

