Identification of factors regulating poly(A) tail synthesis and maturation
- PMID: 15121841
- PMCID: PMC400472
- DOI: 10.1128/MCB.24.10.4196-4206.2004
Identification of factors regulating poly(A) tail synthesis and maturation
Abstract
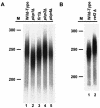
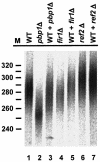
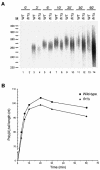
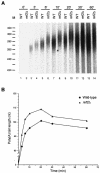
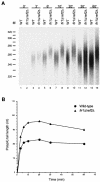
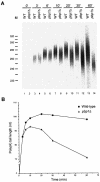
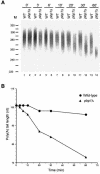
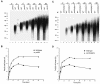
Posttranscriptional maturation of the 3' end of eukaryotic pre-mRNAs occurs as a three-step pathway involving site-specific cleavage, polymerization of a poly(A) tail, and trimming of the newly synthesized tail to its mature length. While most of the factors essential for catalyzing these reactions have been identified, those that regulate them remain to be characterized. Previously, we demonstrated that the yeast protein Pbp1p associates with poly(A)-binding protein (Pab1p) and controls the extent of mRNA polyadenylation. To further elucidate the function of Pbp1p, we conducted a two-hybrid screen to identify factors with which it interacts. Five genes encoding putative Pbp1p-interacting proteins were identified, including (i) FIR1/PIP1 and UFD1/PIP3, genes encoding factors previously implicated in mRNA 3'-end processing; (ii) PBP1 itself, confirming directed two-hybrid results and suggesting that Pbp1p can multimerize; (iii) DIG1, encoding a mitogen-activated protein kinase-associated protein; and (iv) PBP4 (YDL053C), a previously uncharacterized gene. In vitro polyadenylation reactions utilizing extracts derived from fir1 Delta and pbp1 Delta cells and from cells lacking the Fir1p interactor, Ref2p, demonstrated that Pbp1p, Fir1p, and Ref2p are all required for the formation of a normal-length poly(A) tail on precleaved CYC1 pre-mRNA. Kinetic analyses of the respective polyadenylation reactions indicated that Pbp1p is a negative regulator of poly(A) nuclease (PAN) activity and that Fir1p and Ref2p are, respectively, a positive regulator and a negative regulator of poly(A) synthesis. We suggest a model in which these three factors and Ufd1p are part of a regulatory complex that exploits Pab1p to link cleavage and polyadenylation factors of CFIA and CFIB (cleavage factors IA and IB) to the polyadenylation factors of CPF (cleavage and polyadenylation factor).
Figures

Similar articles
-
Pbp1p, a factor interacting with Saccharomyces cerevisiae poly(A)-binding protein, regulates polyadenylation.Mol Cell Biol. 1998 Dec;18(12):7383-96. doi: 10.1128/MCB.18.12.7383. Mol Cell Biol. 1998. PMID: 9819425 Free PMC article.
-
Positive and negative regulation of poly(A) nuclease.Mol Cell Biol. 2004 Jun;24(12):5521-33. doi: 10.1128/MCB.24.12.5521-5533.2004. Mol Cell Biol. 2004. PMID: 15169912 Free PMC article.
-
Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA.Mol Cell Biol. 1992 Aug;12(8):3470-81. doi: 10.1128/mcb.12.8.3470-3481.1992. Mol Cell Biol. 1992. PMID: 1352851 Free PMC article.
-
A history of poly A sequences: from formation to factors to function.Prog Nucleic Acid Res Mol Biol. 2002;71:285-389. doi: 10.1016/s0079-6603(02)71046-5. Prog Nucleic Acid Res Mol Biol. 2002. PMID: 12102557 Review.
-
Birth of a poly(A) tail: mechanisms and control of mRNA polyadenylation.FEBS Open Bio. 2023 Jul;13(7):1140-1153. doi: 10.1002/2211-5463.13528. Epub 2022 Dec 7. FEBS Open Bio. 2023. PMID: 36416579 Free PMC article. Review.
Cited by
-
Initiation of the TORC1-regulated G0 program requires Igo1/2, which license specific mRNAs to evade degradation via the 5'-3' mRNA decay pathway.Mol Cell. 2010 May 14;38(3):345-55. doi: 10.1016/j.molcel.2010.02.039. Mol Cell. 2010. PMID: 20471941 Free PMC article.
-
Pbp1-Interacting Protein Mkt1 Regulates Virulence and Sexual Reproduction in Cryptococcus neoformans.Front Cell Infect Microbiol. 2019 Oct 17;9:355. doi: 10.3389/fcimb.2019.00355. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31681631 Free PMC article.
-
Multiple RNAs from the mouse carboxypeptidase M locus: functional RNAs or transcription noise?BMC Mol Biol. 2009 Feb 8;10:7. doi: 10.1186/1471-2199-10-7. BMC Mol Biol. 2009. PMID: 19200403 Free PMC article.
-
Trypanosome MKT1 and the RNA-binding protein ZC3H11: interactions and potential roles in post-transcriptional regulatory networks.Nucleic Acids Res. 2014 Apr;42(7):4652-68. doi: 10.1093/nar/gkt1416. Epub 2014 Jan 26. Nucleic Acids Res. 2014. PMID: 24470144 Free PMC article.
-
Ataxin-2: From RNA Control to Human Health and Disease.Genes (Basel). 2017 Jun 5;8(6):157. doi: 10.3390/genes8060157. Genes (Basel). 2017. PMID: 28587229 Free PMC article. Review.
References
-
- Amrani, N., M. E. Dufour, N. Bonneaud, and F. Lacroute. 1996. Mutations in STS1 suppress the defect in 3′ mRNA processing caused by the rna15-2 mutation in Saccharomyces cerevisiae. Mol. Gen. Genet. 252:552-562. - PubMed
-
- Bartel, P., C. T. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14:920-924. - PubMed
-
- Bartel, P. L., and S. Fields. 1997. The yeast two-hybrid system. Oxford University Press, New York, N.Y.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
