Multidimensional signatures in antimicrobial peptides
- PMID: 15118082
- PMCID: PMC409924
- DOI: 10.1073/pnas.0401567101
Multidimensional signatures in antimicrobial peptides
Abstract
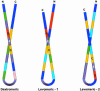
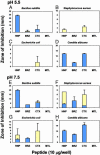
Conventional analyses distinguish between antimicrobial peptides by differences in amino acid sequence. Yet structural paradigms common to broader classes of these molecules have not been established. The current analyses examined the potential conservation of structural themes in antimicrobial peptides from evolutionarily diverse organisms. Using proteomics, an antimicrobial peptide signature was discovered to integrate stereospecific sequence patterns and a hallmark three-dimensional motif. This striking multidimensional signature is conserved among disulfide-containing antimicrobial peptides spanning biological kingdoms, and it transcends motifs previously limited to defined peptide subclasses. Experimental data validating this model enabled the identification of previously unrecognized antimicrobial activity in peptides of known identity. The multidimensional signature model provides a unifying structural theme in broad classes of antimicrobial peptides, will facilitate discovery of antimicrobial peptides as yet unknown, and offers insights into the evolution of molecular determinants in these and related host defense effector molecules.
Figures




Similar articles
-
Structural congruence among membrane-active host defense polypeptides of diverse phylogeny.Biochim Biophys Acta. 2006 Sep;1758(9):1373-86. doi: 10.1016/j.bbamem.2006.03.027. Epub 2006 Apr 19. Biochim Biophys Acta. 2006. PMID: 16725105
-
Structural correlates of antimicrobial efficacy in IL-8 and related human kinocidins.Biochim Biophys Acta. 2007 Mar;1768(3):598-608. doi: 10.1016/j.bbamem.2006.11.011. Epub 2006 Nov 30. Biochim Biophys Acta. 2007. PMID: 17208195
-
The gamma-core motif correlates with antimicrobial activity in cysteine-containing kaliocin-1 originating from transferrins.Biochim Biophys Acta. 2007 Nov;1768(11):2862-72. doi: 10.1016/j.bbamem.2007.07.024. Epub 2007 Aug 16. Biochim Biophys Acta. 2007. PMID: 17916323
-
Unifying themes in host defence effector polypeptides.Nat Rev Microbiol. 2007 Sep;5(9):727-40. doi: 10.1038/nrmicro1744. Nat Rev Microbiol. 2007. PMID: 17703227 Review.
-
Cysteine-rich antimicrobial peptides in invertebrates.Biopolymers. 1998;47(6):465-77. doi: 10.1002/(SICI)1097-0282(1998)47:6<465::AID-BIP5>3.0.CO;2-#. Biopolymers. 1998. PMID: 10333738 Review.
Cited by
-
Properties and mechanisms of action of naturally occurring antifungal peptides.Cell Mol Life Sci. 2013 Oct;70(19):3545-70. doi: 10.1007/s00018-013-1260-1. Epub 2013 Feb 5. Cell Mol Life Sci. 2013. PMID: 23381653 Free PMC article. Review.
-
Identification of defensin-encoding genes of Picea glauca: characterization of PgD5, a conserved spruce defensin with strong antifungal activity.BMC Plant Biol. 2012 Oct 5;12:180. doi: 10.1186/1471-2229-12-180. BMC Plant Biol. 2012. PMID: 23035776 Free PMC article.
-
Beta-sheet pore-forming peptides selected from a rational combinatorial library: mechanism of pore formation in lipid vesicles and activity in biological membranes.Biochemistry. 2007 Oct 30;46(43):12124-39. doi: 10.1021/bi700978h. Epub 2007 Oct 6. Biochemistry. 2007. PMID: 17918962 Free PMC article.
-
A review of antimicrobial peptides and their therapeutic potential as anti-infective drugs.Curr Eye Res. 2005 Jul;30(7):505-15. doi: 10.1080/02713680590968637. Curr Eye Res. 2005. PMID: 16020284 Free PMC article. Review.
-
Structure--activity study of the antibacterial peptide fallaxin.Protein Sci. 2007 Sep;16(9):1969-76. doi: 10.1110/ps.072966007. Protein Sci. 2007. PMID: 17766389 Free PMC article.
References
-
- Yeaman, M. R. & Yount, N. Y. (2003) Pharmacol. Rev. 55, 27-55. - PubMed
-
- Blaser, M. J. (2002) N. Engl. J. Med. 346, 2083-2085. - PubMed
-
- White, S. H., Wimley, W. C. & Selsted, M. E. (1995) Curr. Opin. Struct. Biol. 5, 521-527. - PubMed
-
- Guder, A., Wiedemann, I. & Sahl, H. G. (2000) Biopolymers 55, 62-73. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

