Structure and mechanism of the RNA polymerase II transcription machinery
- PMID: 15114340
- PMCID: PMC1189732
- DOI: 10.1038/nsmb763
Structure and mechanism of the RNA polymerase II transcription machinery
Abstract
Advances in structure determination of the bacterial and eukaryotic transcription machinery have led to a marked increase in the understanding of the mechanism of transcription. Models for the specific assembly of the RNA polymerase II transcription machinery at a promoter, conformational changes that occur during initiation of transcription, and the mechanism of initiation are discussed in light of recent developments.
Conflict of interest statement
The author declares that he has no competing financial interests.
Figures
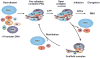
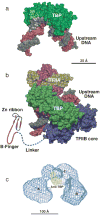
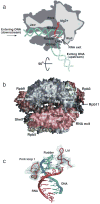
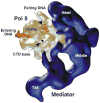

Similar articles
-
Structural basis of transcription: separation of RNA from DNA by RNA polymerase II.Science. 2004 Feb 13;303(5660):1014-6. doi: 10.1126/science.1090839. Science. 2004. PMID: 14963331
-
The RNA polymerase II core promoter.Annu Rev Biochem. 2003;72:449-79. doi: 10.1146/annurev.biochem.72.121801.161520. Epub 2003 Mar 19. Annu Rev Biochem. 2003. PMID: 12651739 Review.
-
Eukaryotic transcription initiation machinery visualized at molecular level.Transcription. 2016 Oct 19;7(5):203-208. doi: 10.1080/21541264.2016.1237150. Transcription. 2016. PMID: 27658022 Free PMC article. Review.
-
RNA polymerase II as a control panel for multiple coactivator complexes.Curr Opin Genet Dev. 1999 Apr;9(2):132-9. doi: 10.1016/S0959-437X(99)80020-3. Curr Opin Genet Dev. 1999. PMID: 10322136 Review.
-
Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 Angstroms.Science. 2004 Feb 13;303(5660):983-8. doi: 10.1126/science.1090838. Science. 2004. PMID: 14963322
Cited by
-
Regulation of mammalian transcription by Gdown1 through a novel steric crosstalk revealed by cryo-EM.EMBO J. 2012 Aug 29;31(17):3575-87. doi: 10.1038/emboj.2012.205. Epub 2012 Jul 31. EMBO J. 2012. PMID: 22850672 Free PMC article.
-
Inactivated RNA polymerase II open complexes can be reactivated with TFIIE.J Biol Chem. 2012 Jan 6;287(2):961-7. doi: 10.1074/jbc.M111.297572. Epub 2011 Nov 27. J Biol Chem. 2012. PMID: 22119917 Free PMC article.
-
Rethinking the role of TFIIF in transcript initiation by RNA polymerase II.Transcription. 2012 Jul-Aug;3(4):156-9. doi: 10.4161/trns.20725. Epub 2012 Jul 1. Transcription. 2012. PMID: 22771986 Free PMC article. Review.
-
The emerging role of RNA polymerase I transcription machinery in human malignancy: a clinical perspective.Onco Targets Ther. 2013 Jul 19;6:909-16. doi: 10.2147/OTT.S36627. Print 2013. Onco Targets Ther. 2013. PMID: 23888116 Free PMC article.
-
Nuclear import of TFIIB is mediated by Kap114p, a karyopherin with multiple cargo-binding domains.Mol Biol Cell. 2005 Jul;16(7):3200-10. doi: 10.1091/mbc.e04-11-0990. Epub 2005 May 11. Mol Biol Cell. 2005. PMID: 15888545 Free PMC article.
References
-
- Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77–137. - PubMed
-
- Woychik NA, Hampsey M. The RNA polymerase II machinery: structure illuminates function. Cell. 2002;108:453–463. - PubMed
-
- Borukhov S, Nudler E. RNA polymerase holoenzyme: structure, function and biological implications. Curr Opin Microbiol. 2003;6:93–100. - PubMed
-
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

