Accumulation of a 3'-terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells
- PMID: 15113895
- PMCID: PMC400339
- DOI: 10.1128/jvi.78.10.5133-5138.2004
Accumulation of a 3'-terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells
Abstract
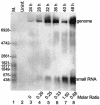
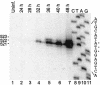
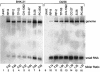
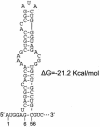
Japanese encephalitis virus (JEV) contains a single positive-strand RNA genome nearly 11 kb in length and is not formally thought to generate subgenomic RNA molecules during replication. Here, we report the abundant accumulation of a 3'-terminal 521- to 523-nucleotide (nt) genome fragment, representing a major portion of the 585-nt 3' untranslated region, in both mammalian (BHK-21) and mosquito (C6/36) cells infected with any of nine strains of JEV. In BHK-21 cells, the viral genome was detected as early as 24 h postinfection, the small RNA was detected as early as 28 h postinfection, and the small RNA was 0.25 to 1.5 times as abundant as the genome on a molar basis between 28 and 48 h postinfection. In C6/36 cells, the genome and small RNA were present 5 days postinfection and the small RNA was 1.25 to 5.14 times as abundant as the genome. The 3'-terminal 523-nt small RNA contains a 5'-proximal stable hairpin (nt 6 to 56) that may play a role in its formation and the conserved flavivirus 3'-cyclization motif (nt 413 to 420) and the 3'-terminal long stable hairpin structure (nt 440 to 523) that have postulated roles in genome replication. Abundant accumulation of the small RNA during viral replication in both mammalian and mosquito cells suggests that it may play a biological role, perhaps as a regulator of RNA synthesis.
Figures
Similar articles
-
The conserved stem-loop II structure at the 3' untranslated region of Japanese encephalitis virus genome is required for the formation of subgenomic flaviviral RNA.PLoS One. 2018 Jul 26;13(7):e0201250. doi: 10.1371/journal.pone.0201250. eCollection 2018. PLoS One. 2018. PMID: 30048535 Free PMC article.
-
Defective interfering RNAs of Japanese encephalitis virus found in mosquito cells and correlation with persistent infection.Virus Res. 2007 Mar;124(1-2):139-50. doi: 10.1016/j.virusres.2006.10.013. Epub 2006 Nov 28. Virus Res. 2007. PMID: 17134784
-
3' cis-acting elements that contribute to the competence and efficiency of Japanese encephalitis virus genome replication: functional importance of sequence duplications, deletions, and substitutions.J Virol. 2009 Aug;83(16):7909-30. doi: 10.1128/JVI.02541-08. Epub 2009 Jun 3. J Virol. 2009. PMID: 19494005 Free PMC article.
-
Integrative effect of defective interfering RNA accumulation and helper virus attenuation is responsible for the persistent infection of Japanese encephalitis virus in BHK-21 cells.J Med Virol. 2013 Nov;85(11):1990-2000. doi: 10.1002/jmv.23665. Epub 2013 Jul 16. J Med Virol. 2013. PMID: 23861255
-
The A66G back mutation in NS2A of JEV SA14-14-2 strain contributes to production of NS1' protein and the secreted NS1' can be used for diagnostic biomarker for virulent virus infection.Infect Genet Evol. 2015 Dec;36:116-125. doi: 10.1016/j.meegid.2015.09.013. Epub 2015 Sep 16. Infect Genet Evol. 2015. PMID: 26384477
Cited by
-
Small noncoding RNA modulates Japanese encephalitis virus replication and translation in trans.Virol J. 2011 Nov 1;8:492. doi: 10.1186/1743-422X-8-492. Virol J. 2011. PMID: 22040380 Free PMC article.
-
West Nile virus infection does not induce PKR activation in rodent cells.Virology. 2011 Dec 5;421(1):51-60. doi: 10.1016/j.virol.2011.08.008. Epub 2011 Oct 7. Virology. 2011. PMID: 21982595 Free PMC article.
-
Viral long non-coding RNA regulates virus life-cycle and pathogenicity.Mol Biol Rep. 2022 Jul;49(7):6693-6700. doi: 10.1007/s11033-022-07268-6. Epub 2022 Mar 17. Mol Biol Rep. 2022. PMID: 35301646 Free PMC article. Review.
-
The conserved stem-loop II structure at the 3' untranslated region of Japanese encephalitis virus genome is required for the formation of subgenomic flaviviral RNA.PLoS One. 2018 Jul 26;13(7):e0201250. doi: 10.1371/journal.pone.0201250. eCollection 2018. PLoS One. 2018. PMID: 30048535 Free PMC article.
-
A viral noncoding RNA generated by cis-element-mediated protection against 5'->3' RNA decay represses both cap-independent and cap-dependent translation.J Virol. 2008 Oct;82(20):10162-74. doi: 10.1128/JVI.01027-08. Epub 2008 Aug 13. J Virol. 2008. PMID: 18701589 Free PMC article.
References
-
- Brinton, M. A. 1982. Characterization of West Nile virus persistent infections in genetically resistant and susceptible mouse cells. I. Generation of defective nonplaquing virus particles. Virology 116:84-98. - PubMed
-
- Brinton, M. A. 1986. Replication of flaviviruses, p. 327-374. In S. Schlesinger and M. J. Schlesinger (ed.), The Togaviridae and Flaviviridae. Plenum Press, Inc., New York, N.Y.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

