Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes
- PMID: 15103327
- PMCID: PMC424369
- DOI: 10.1038/sj.emboj.7600210
Analysis of the composition, assembly kinetics and activity of native Apaf-1 apoptosomes
Abstract
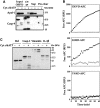
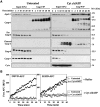
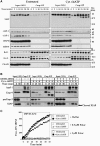
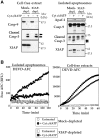
The Apaf-1 apoptosome is a multi-subunit caspase-activating scaffold that is assembled in response to diverse forms of cellular stress that culminate in apoptosis. To date, most studies on apoptosome composition and function have used apoptosomes reassembled from recombinant or purified proteins. Thus, the precise composition of native apoptosomes remains unresolved. Here, we have used a one-step immunopurification approach to isolate catalytically active Apaf-1/caspase-9 apoptosomes, and have identified the major constituents of these complexes using mass spectrometry methods. Using this approach, we have also assessed the ability of putative apoptosome regulatory proteins, such as Smac/DIABLO and PHAPI, to regulate the activity of native apoptosomes. We show that Apaf-1, caspase-9, caspase-3 and XIAP are the major constituents of native apoptosomes and that cytochrome c is not stably associated with the active complex. We also demonstrate that the IAP-neutralizing protein Smac/DIABLO and the tumor-suppressor protein PHAPI can enhance the catalytic activity of apoptosome complexes in distinct ways. Surprisingly, PHAPI also enhanced the activity of purified caspase-3, suggesting that it may act as a co-factor for this protease.
Figures



Similar articles
-
Pro-apoptotic proteins released from the mitochondria regulate the protein composition and caspase-processing activity of the native Apaf-1/caspase-9 apoptosome complex.J Biol Chem. 2004 May 7;279(19):19665-82. doi: 10.1074/jbc.M311388200. Epub 2004 Mar 1. J Biol Chem. 2004. PMID: 14993223
-
XIAP inhibition of caspase-3 preserves its association with the Apaf-1 apoptosome and prevents CD95- and Bax-induced apoptosis.Cell Death Differ. 2002 Sep;9(9):881-92. doi: 10.1038/sj.cdd.4401069. Cell Death Differ. 2002. PMID: 12181739
-
PHAPI, CAS, and Hsp70 promote apoptosome formation by preventing Apaf-1 aggregation and enhancing nucleotide exchange on Apaf-1.Mol Cell. 2008 Apr 25;30(2):239-47. doi: 10.1016/j.molcel.2008.03.014. Mol Cell. 2008. PMID: 18439902
-
Chemical-induced apoptosis: formation of the Apaf-1 apoptosome.Drug Metab Rev. 2003 Nov;35(4):337-63. doi: 10.1081/dmr-120026497. Drug Metab Rev. 2003. PMID: 14705865 Review.
-
Apaf-1: Regulation and function in cell death.Biochimie. 2017 Apr;135:111-125. doi: 10.1016/j.biochi.2017.02.001. Epub 2017 Feb 9. Biochimie. 2017. PMID: 28192157 Review.
Cited by
-
Major pathways of polymyxin-induced apoptosis in rat kidney proximal tubular cells.Antimicrob Agents Chemother. 2015 Apr;59(4):2136-43. doi: 10.1128/AAC.04869-14. Epub 2015 Jan 26. Antimicrob Agents Chemother. 2015. PMID: 25624331 Free PMC article.
-
Differential expression of Fas family members and Bcl-2 family members in benign versus malignant epithelial ovarian cancer (EOC) in North Indian population.Mol Cell Biochem. 2012 Sep;368(1-2):119-26. doi: 10.1007/s11010-012-1350-7. Epub 2012 Jun 12. Mol Cell Biochem. 2012. PMID: 22688594 Clinical Trial.
-
MUC1 oncoprotein blocks nuclear targeting of c-Abl in the apoptotic response to DNA damage.EMBO J. 2006 Aug 23;25(16):3774-83. doi: 10.1038/sj.emboj.7601263. Epub 2006 Aug 3. EMBO J. 2006. PMID: 16888623 Free PMC article.
-
Cell Survival and Cell Death at the Intersection of Autophagy and Apoptosis: Implications for Current and Future Cancer Therapeutics.ACS Pharmacol Transl Sci. 2021 Nov 3;4(6):1728-1746. doi: 10.1021/acsptsci.1c00130. eCollection 2021 Dec 10. ACS Pharmacol Transl Sci. 2021. PMID: 34927007 Free PMC article. Review.
-
Apoptosis in post-streptococcal glomerulonephritis and mechanisms for failed of inflammation resolution.Pediatr Nephrol. 2024 Jun;39(6):1709-1724. doi: 10.1007/s00467-023-06162-y. Epub 2023 Sep 29. Pediatr Nephrol. 2024. PMID: 37775580 Review.
References
-
- Acehan D, Jiang X, Morgan DG, Heuser JE, Wang X, Akey CW (2002) Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Mol Cell 9: 423–432 - PubMed
-
- Adrain C, Slee EA, Harte MT, Martin SJ (1999) Regulation of apoptotic protease activating factor-1 oligomerization and apoptosis by the WD-40 repeat region. J Biol Chem 274: 20855–20860 - PubMed
-
- Beere HM, Wolf BB, Cain K, Mosser DD, Mahboubi A, Kuwana T, Tailor P, Morimoto RI, Cohen GM, Green DR (2000) Heat-shock protein 70 inhibits apoptosis by preventing recruitment of procaspase-9 to the Apaf-1 apoptosome. Nat Cell Biol 2: 469–475 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

