Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor
- PMID: 15096522
- PMCID: PMC2172034
- DOI: 10.1083/jcb.200403022
Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor
Abstract
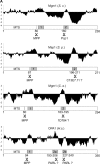
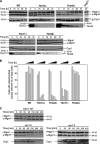
Mitochondrial morphology and inheritance of mitochondrial DNA in yeast depend on the dynamin-like GTPase Mgm1. It is present in two isoforms in the intermembrane space of mitochondria both of which are required for Mgm1 function. Limited proteolysis of the large isoform by the mitochondrial rhomboid protease Pcp1/Rbd1 generates the short isoform of Mgm1 but how this is regulated is unclear. We show that near its NH2 terminus Mgm1 contains two conserved hydrophobic segments of which the more COOH-terminal one is cleaved by Pcp1. Changing the hydrophobicity of the NH2-terminal segment modulated the ratio of the isoforms and led to fragmentation of mitochondria. Formation of the short isoform of Mgm1 and mitochondrial morphology further depend on a functional protein import motor and on the ATP level in the matrix. Our data show that a novel pathway, to which we refer as alternative topogenesis, represents a key regulatory mechanism ensuring the balanced formation of both Mgm1 isoforms. Through this process the mitochondrial ATP level might control mitochondrial morphology.
Figures





Similar articles
-
Intramembrane proteolysis of Mgm1 by the mitochondrial rhomboid protease is highly promiscuous regarding the sequence of the cleaved hydrophobic segment.J Mol Biol. 2010 Aug 13;401(2):182-93. doi: 10.1016/j.jmb.2010.06.014. Epub 2010 Jun 15. J Mol Biol. 2010. PMID: 20558178
-
Distinct roles of the two isoforms of the dynamin-like GTPase Mgm1 in mitochondrial fusion.FEBS Lett. 2009 Jul 7;583(13):2237-43. doi: 10.1016/j.febslet.2009.05.053. Epub 2009 Jun 6. FEBS Lett. 2009. PMID: 19505460
-
Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA.J Biol Chem. 2003 Jul 25;278(30):27781-8. doi: 10.1074/jbc.M211311200. Epub 2003 Apr 21. J Biol Chem. 2003. PMID: 12707284
-
Transport of adenine nucleotides in the mitochondria of Saccharomyces cerevisiae: interactions between the ADP/ATP carriers and the ATP-Mg/Pi carrier.Mitochondrion. 2009 Apr;9(2):79-85. doi: 10.1016/j.mito.2009.01.001. Epub 2009 Jan 17. Mitochondrion. 2009. PMID: 19460304 Review.
-
The protein import motor of mitochondria.Nat Rev Mol Cell Biol. 2002 Aug;3(8):555-65. doi: 10.1038/nrm878. Nat Rev Mol Cell Biol. 2002. PMID: 12154367 Review.
Cited by
-
Mitochondrial dynamics involves molecular and mechanical events in motility, fusion and fission.Front Cell Dev Biol. 2022 Oct 19;10:1010232. doi: 10.3389/fcell.2022.1010232. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36340034 Free PMC article. Review.
-
Mitochondrial Processing Peptidases-Structure, Function and the Role in Human Diseases.Int J Mol Sci. 2022 Jan 24;23(3):1297. doi: 10.3390/ijms23031297. Int J Mol Sci. 2022. PMID: 35163221 Free PMC article. Review.
-
Pathways shaping the mitochondrial inner membrane.Open Biol. 2021 Dec;11(12):210238. doi: 10.1098/rsob.210238. Epub 2021 Dec 1. Open Biol. 2021. PMID: 34847778 Free PMC article. Review.
-
The mitochondrial intermembrane space: the most constricted mitochondrial sub-compartment with the largest variety of protein import pathways.Open Biol. 2021 Mar;11(3):210002. doi: 10.1098/rsob.210002. Epub 2021 Mar 10. Open Biol. 2021. PMID: 33715390 Free PMC article. Review.
-
Inhibition of the mitochondrial ATPase function by IF1 changes the spatiotemporal organization of ATP synthase.Biochim Biophys Acta Bioenerg. 2021 Jan 1;1862(1):148322. doi: 10.1016/j.bbabio.2020.148322. Epub 2020 Oct 14. Biochim Biophys Acta Bioenerg. 2021. PMID: 33065099 Free PMC article.
References
-
- Alexander, C., M. Votruba, U.E. Pesch, D.L. Thiselton, S. Mayer, A. Moore, M. Rodriguez, U. Kellner, B. Leo-Kottler, G. Auburger, et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 26:211–215. - PubMed
-
- Delettre, C., G. Lenaers, J.M. Griffoin, N. Gigarel, C. Lorenzo, P. Belenguer, L. Pelloquin, J. Grosgeorge, C. Turc-Carel, E. Perret, et al. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy. Nat. Genet. 26:207–210. - PubMed
-
- Esser, K., B. Tursun, M. Ingenhoven, G. Michaelis, and E. Pratje. 2002. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease pcp1. J. Mol. Biol. 323:835–843. - PubMed
-
- Foury, F., and A. Tzagoloff. 1976. Localization on mitochondrial DNA of mutations leading to a loss of rutamycin-sensitive adenosine triphosphatase. Eur. J. Biochem. 68:113–119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

