Predicting co-complexed protein pairs using genomic and proteomic data integration
- PMID: 15090078
- PMCID: PMC419405
- DOI: 10.1186/1471-2105-5-38
Predicting co-complexed protein pairs using genomic and proteomic data integration
Abstract
Background: Identifying all protein-protein interactions in an organism is a major objective of proteomics. A related goal is to know which protein pairs are present in the same protein complex. High-throughput methods such as yeast two-hybrid (Y2H) and affinity purification coupled with mass spectrometry (APMS) have been used to detect interacting proteins on a genomic scale. However, both Y2H and APMS methods have substantial false-positive rates. Aside from high-throughput interaction screens, other gene- or protein-pair characteristics may also be informative of physical interaction. Therefore it is desirable to integrate multiple datasets and utilize their different predictive value for more accurate prediction of co-complexed relationship.
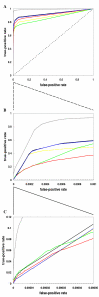
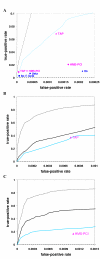
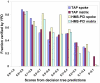
Results: Using a supervised machine learning approach--probabilistic decision tree, we integrated high-throughput protein interaction datasets and other gene- and protein-pair characteristics to predict co-complexed pairs (CCP) of proteins. Our predictions proved more sensitive and specific than predictions based on Y2H or APMS methods alone or in combination. Among the top predictions not annotated as CCPs in our reference set (obtained from the MIPS complex catalogue), a significant fraction was found to physically interact according to a separate database (YPD, Yeast Proteome Database), and the remaining predictions may potentially represent unknown CCPs.
Conclusions: We demonstrated that the probabilistic decision tree approach can be successfully used to predict co-complexed protein (CCP) pairs from other characteristics. Our top-scoring CCP predictions provide testable hypotheses for experimental validation.
Figures

Similar articles
-
A computational approach for ordering signal transduction pathway components from genomics and proteomics Data.BMC Bioinformatics. 2004 Oct 25;5:158. doi: 10.1186/1471-2105-5-158. BMC Bioinformatics. 2004. PMID: 15504238 Free PMC article.
-
AVID: an integrative framework for discovering functional relationships among proteins.BMC Bioinformatics. 2005 Jun 1;6:136. doi: 10.1186/1471-2105-6-136. BMC Bioinformatics. 2005. PMID: 15929793 Free PMC article.
-
A high-accuracy consensus map of yeast protein complexes reveals modular nature of gene essentiality.BMC Bioinformatics. 2007 Jul 2;8:236. doi: 10.1186/1471-2105-8-236. BMC Bioinformatics. 2007. PMID: 17605818 Free PMC article.
-
Computational detection of protein complexes in AP-MS experiments.Proteomics. 2012 May;12(10):1663-8. doi: 10.1002/pmic.201100508. Proteomics. 2012. PMID: 22711593 Review.
-
Functional genomics and proteomics in the clinical neurosciences: data mining and bioinformatics.Prog Brain Res. 2006;158:83-108. doi: 10.1016/S0079-6123(06)58004-5. Prog Brain Res. 2006. PMID: 17027692 Review.
Cited by
-
Protein-protein interaction prediction using a hybrid feature representation and a stacked generalization scheme.BMC Bioinformatics. 2019 Jun 10;20(1):308. doi: 10.1186/s12859-019-2907-1. BMC Bioinformatics. 2019. PMID: 31182027 Free PMC article.
-
Bayesian inference for genomic data integration reduces misclassification rate in predicting protein-protein interactions.PLoS Comput Biol. 2011 Jul;7(7):e1002110. doi: 10.1371/journal.pcbi.1002110. Epub 2011 Jul 28. PLoS Comput Biol. 2011. PMID: 21829334 Free PMC article.
-
Protein complex identification by integrating protein-protein interaction evidence from multiple sources.PLoS One. 2013 Dec 27;8(12):e83841. doi: 10.1371/journal.pone.0083841. eCollection 2013. PLoS One. 2013. PMID: 24386289 Free PMC article.
-
The development of a universal in silico predictor of protein-protein interactions.PLoS One. 2013 May 31;8(5):e65587. doi: 10.1371/journal.pone.0065587. Print 2013. PLoS One. 2013. PMID: 23741499 Free PMC article.
-
A novel biclustering approach to association rule mining for predicting HIV-1-human protein interactions.PLoS One. 2012;7(4):e32289. doi: 10.1371/journal.pone.0032289. Epub 2012 Apr 23. PLoS One. 2012. PMID: 22539940 Free PMC article.
References
-
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. - DOI - PubMed
-
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Toward a protein-protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci U S A. 2000;97:1143–1147. doi: 10.1073/pnas.97.3.1143. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

