The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis
- PMID: 15087503
- PMCID: PMC404069
- DOI: 10.1073/pnas.0308686101
The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis
Abstract
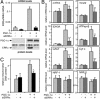
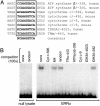
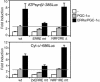
Estrogen-related receptor alpha (ERRalpha) is one of the first orphan nuclear receptors to be identified, yet its physiological functions are still unclear. We show here that ERRalpha is an effector of the transcriptional coactivator PGC-1alpha [peroxisome proliferator-activated receptor gamma (PPARgamma) coactivator 1alpha], and that it regulates the expression of genes involved in oxidative phosphorylation and mitochondrial biogenesis. Inhibition of ERRalpha compromises the ability of PGC-1alpha to induce the expression of genes encoding mitochondrial proteins and to increase mitochondrial DNA content. A constitutively active form of ERRalpha is sufficient to elicit both responses. ERRalpha binding sites are present in the transcriptional control regions of ERRalpha/PGC-1alpha-induced genes and contribute to the transcriptional response to PGC-1alpha. The ERRalpha-regulated genes described here have been reported to be expressed at reduced levels in humans that are insulin-resistant. Thus, changes in ERRalpha activity could be linked to pathological changes in metabolic disease, such as diabetes.
Figures


Similar articles
-
Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle.Proc Natl Acad Sci U S A. 2004 Apr 27;101(17):6570-5. doi: 10.1073/pnas.0401401101. Epub 2004 Apr 20. Proc Natl Acad Sci U S A. 2004. PMID: 15100410 Free PMC article.
-
Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand.Proc Natl Acad Sci U S A. 2004 Jun 15;101(24):8912-7. doi: 10.1073/pnas.0401420101. Epub 2004 Jun 7. Proc Natl Acad Sci U S A. 2004. PMID: 15184675 Free PMC article.
-
Estrogen-related receptor alpha directs peroxisome proliferator-activated receptor alpha signaling in the transcriptional control of energy metabolism in cardiac and skeletal muscle.Mol Cell Biol. 2004 Oct;24(20):9079-91. doi: 10.1128/MCB.24.20.9079-9091.2004. Mol Cell Biol. 2004. PMID: 15456881 Free PMC article.
-
[Transcriptional regulation of metabolic switching PDK4 gene under various physiological conditions].Yakugaku Zasshi. 2007 Jan;127(1):153-62. doi: 10.1248/yakushi.127.153. Yakugaku Zasshi. 2007. PMID: 17202796 Review. Japanese.
-
Natural products, PGC-1 α , and Duchenne muscular dystrophy.Acta Pharm Sin B. 2020 May;10(5):734-745. doi: 10.1016/j.apsb.2020.01.001. Epub 2020 Jan 8. Acta Pharm Sin B. 2020. PMID: 32528825 Free PMC article. Review.
Cited by
-
Adaptability to hypobaric hypoxia is facilitated through mitochondrial bioenergetics: an in vivo study.Br J Pharmacol. 2013 Jul;169(5):1035-47. doi: 10.1111/bph.12179. Br J Pharmacol. 2013. PMID: 23517027 Free PMC article.
-
PGC1α expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress.Cancer Cell. 2013 Mar 18;23(3):287-301. doi: 10.1016/j.ccr.2012.11.020. Epub 2013 Feb 14. Cancer Cell. 2013. PMID: 23416000 Free PMC article.
-
Estrogen-related receptor α in normal adrenal cortex and adrenocortical tumors: involvement in development and oncogenesis.Mol Cell Endocrinol. 2013 Jan 30;365(2):207-11. doi: 10.1016/j.mce.2012.10.020. Epub 2012 Oct 30. Mol Cell Endocrinol. 2013. PMID: 23123734 Free PMC article.
-
NT-PGC-1α protein is sufficient to link β3-adrenergic receptor activation to transcriptional and physiological components of adaptive thermogenesis.J Biol Chem. 2012 Mar 16;287(12):9100-11. doi: 10.1074/jbc.M111.320200. Epub 2012 Jan 26. J Biol Chem. 2012. PMID: 22282499 Free PMC article.
-
The diverse role of the PPARγ coactivator 1 family of transcriptional coactivators in cancer.Semin Cell Dev Biol. 2012 Jun;23(4):381-8. doi: 10.1016/j.semcdb.2012.01.007. Epub 2012 Jan 21. Semin Cell Dev Biol. 2012. PMID: 22285815 Free PMC article. Review.
References
-
- Giguere, V., Yang, N., Segui, P. & Evans, R. M. (1988) Nature 331, 91-94. - PubMed
-
- Yang, N., Shigeta, H., Shi, H. & Teng, C. T. (1996) J. Biol. Chem. 271, 5795-5804. - PubMed
-
- Johnston, S. D., Liu, X., Zuo, F., Eisenbraun, T. L., Wiley, S. R., Kraus, R. J. & Mertz, J. E. (1997) Mol. Endocrinol. 11, 342-352. - PubMed
-
- Giguere, V. (2002) Trends Endocrinol. Metab. 13, 220-225. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

