The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication
- PMID: 15082529
- PMCID: PMC387417
- DOI: 10.1101/gad.1173204
The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication
Abstract
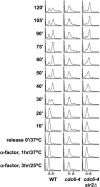
The establishment of DNA synthesis during the S phase is a multistep process that occurs in several stages beginning in late mitosis. The first step is the formation of a large prereplicative complex (pre-RC) at individual replication origins and occurs during exit from mitosis and entry into G1 phase. To better understand the genetic requirements for pre-RC formation, we selected chromosomal suppressors of a temperature-sensitive cdc6-4 mutant defective for pre-RC assembly. Loss-of-function mutations in the chromatin-modifying genes SIR2, and to a lesser extent in SIR3 and SIR4, suppressed the cdc6-4 temperature-sensitive lethality. This suppression was independent of the well-known silencing roles for the SIR proteins at the HM loci, at telomeres, or at the rDNA locus. A deletion of SIR2 uniquely rescued both the DNA synthesis defect of the cdc6-4 mutant and its severe plasmid instability phenotype for many origins. A SIR2 deletion suppressed additional initiation mutants affecting pre-RC assembly but not mutants that act subsequently. These findings suggest that Sir2p negatively regulates the initiation of DNA replication through a novel mechanism and reveal another connection between proteins that initiate DNA synthesis and those that establish silent heterochromatin in budding yeast.
Figures

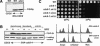

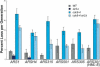

Similar articles
-
An ARS element inhibits DNA replication through a SIR2-dependent mechanism.Mol Cell. 2008 Apr 25;30(2):156-66. doi: 10.1016/j.molcel.2008.02.019. Mol Cell. 2008. PMID: 18439895
-
Locus specificity determinants in the multifunctional yeast silencing protein Sir2.EMBO J. 2000 Jun 1;19(11):2641-51. doi: 10.1093/emboj/19.11.2641. EMBO J. 2000. PMID: 10835361 Free PMC article.
-
Isolation and characterization of conditional alleles of the yeast SIR2 gene.J Mol Biol. 2007 Apr 13;367(5):1246-57. doi: 10.1016/j.jmb.2007.01.044. Epub 2007 Jan 23. J Mol Biol. 2007. PMID: 17316680
-
A model for step-wise assembly of heterochromatin in yeast.Novartis Found Symp. 2004;259:48-56; discussion 56-62, 163-9. Novartis Found Symp. 2004. PMID: 15171246 Review.
-
To fire or not to fire: origin activation in Saccharomyces cerevisiae ribosomal DNA.Genes Dev. 2002 Oct 1;16(19):2459-64. doi: 10.1101/gad.1033702. Genes Dev. 2002. PMID: 12368256 Review. No abstract available.
Cited by
-
The histone acetyltransferases CBP and Chameau integrate developmental and DNA replication programs in Drosophila ovarian follicle cells.Development. 2012 Oct;139(20):3880-90. doi: 10.1242/dev.083576. Epub 2012 Sep 5. Development. 2012. PMID: 22951641 Free PMC article.
-
Nucleotide supply, not local histone acetylation, sets replication origin usage in transcribed regions.EMBO Rep. 2010 Sep;11(9):698-704. doi: 10.1038/embor.2010.112. Epub 2010 Jul 30. EMBO Rep. 2010. PMID: 20671737 Free PMC article.
-
Expanded role for the nitrogen assimilation control protein in the response of Klebsiella pneumoniae to nitrogen stress.J Bacteriol. 2010 Oct;192(19):4812-20. doi: 10.1128/JB.00931-09. Epub 2010 Mar 26. J Bacteriol. 2010. PMID: 20348267 Free PMC article.
-
Linking DNA replication to heterochromatin silencing and epigenetic inheritance.Acta Biochim Biophys Sin (Shanghai). 2012 Jan;44(1):3-13. doi: 10.1093/abbs/gmr107. Acta Biochim Biophys Sin (Shanghai). 2012. PMID: 22194009 Free PMC article. Review.
-
Differential chromatin structure encompassing replication origins in transformed and normal cells.Genes Cancer. 2012 Feb;3(2):152-76. doi: 10.1177/1947601912457026. Genes Cancer. 2012. PMID: 23050047 Free PMC article.
References
-
- Aparicio O.M., Billington, B.L., and Gottschling, D.E. 1991. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell 66: 1279-1287. - PubMed
-
- Aparicio O.M., Weinstein, D.M., and Bell, S.P. 1997. Components and dynamics of DNA replication complexes in S. cerevisiae: Redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59-69. - PubMed
-
- Bell S.P. and Dutta, A. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71: 333-374. - PubMed
-
- Bell S.P. and Stillman, B. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128-134. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
