A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana
- PMID: 15079073
- PMCID: PMC395963
- DOI: 10.1073/pnas.0304346101
A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana
Abstract
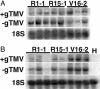
Nicotiana benthamiana often displays more intense symptoms after infection by RNA viruses than do other Nicotiana species. Here, we examined the role of RNA-dependent RNA polymerases (RdRPs) in N. benthamiana antiviral defense. cDNAs representing only two genes encoding RdRPs were identified in N. benthamiana. One RdRP was similar in sequence to SDE1/SGS2 required for maintenance of transgene silencing, whereas the second, named NbRdRP1m, was >90% identical in sequence to the salicylic acid (SA)-inducible RdRP from Nicotiana tabacum required for defense against viruses. NbRdRP1m expression was induced by SA treatment or challenge with Tobacco mosaic virus, but the gene and transcript sequences differed from those of other SA-inducible RdRPs in that they contained a 72-nt insert with tandem in-frame stop codons in the 5' portion of the ORF. N. benthamiana plants transformed with an SA-inducible RdRP gene from Medicago truncatula were more resistant to infection by Tobacco mosaic virus, Turnip vein-clearing virus, and Sunn hemp mosaic virus (members of Tobamovirus genus), but not to Cucumber mosaic virus and Potato virus X (members of different genera than the tobamoviruses). Our results indicate that N. benthamiana lacks an active SA- and virus-inducible RdRP and thus is hypersusceptible to viruses normally limited in their accumulation by this RdRP. These findings are significant for those studying virus-induced gene silencing, the hypersensitive response and systemic acquired resistance.
Figures

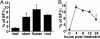
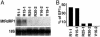


Similar articles
-
Salicylic acid treatment and expression of an RNA-dependent RNA polymerase 1 transgene inhibit lethal symptoms and meristem invasion during tobacco mosaic virus infection in Nicotiana benthamiana.BMC Plant Biol. 2016 Jan 13;16:15. doi: 10.1186/s12870-016-0705-8. BMC Plant Biol. 2016. PMID: 26757721 Free PMC article.
-
RNA-dependent RNA polymerase 1 from Nicotiana tabacum suppresses RNA silencing and enhances viral infection in Nicotiana benthamiana.Plant Cell. 2010 Apr;22(4):1358-72. doi: 10.1105/tpc.109.072058. Epub 2010 Apr 16. Plant Cell. 2010. PMID: 20400679 Free PMC article.
-
Genetic modification of alternative respiration in Nicotiana benthamiana affects basal and salicylic acid-induced resistance to potato virus X.BMC Plant Biol. 2011 Feb 28;11:41. doi: 10.1186/1471-2229-11-41. BMC Plant Biol. 2011. PMID: 21356081 Free PMC article.
-
Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense.Mol Plant Microbe Interact. 2003 Mar;16(3):206-16. doi: 10.1094/MPMI.2003.16.3.206. Mol Plant Microbe Interact. 2003. PMID: 12650452
-
Tobacco RNA-dependent RNA polymerase 1 affects the expression of defence-related genes in Nicotiana benthamiana upon Tomato leaf curl Gujarat virus infection.Planta. 2020 Jul 1;252(1):11. doi: 10.1007/s00425-020-03417-y. Planta. 2020. PMID: 32613448
Cited by
-
Influence of host chloroplast proteins on Tobacco mosaic virus accumulation and intercellular movement.Plant Physiol. 2013 Jan;161(1):134-47. doi: 10.1104/pp.112.207860. Epub 2012 Oct 24. Plant Physiol. 2013. PMID: 23096159 Free PMC article.
-
Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR.PLoS One. 2012;7(9):e46451. doi: 10.1371/journal.pone.0046451. Epub 2012 Sep 28. PLoS One. 2012. PMID: 23029521 Free PMC article.
-
Expressional and regulatory characterization of Arabidopsis RNA-dependent RNA polymerase 1.Planta. 2013 Jun;237(6):1561-9. doi: 10.1007/s00425-013-1863-7. Epub 2013 Mar 16. Planta. 2013. PMID: 23503757
-
Plant viruses. Invaders of cells and pirates of cellular pathways.Plant Physiol. 2005 Aug;138(4):1809-14. doi: 10.1104/pp.104.900167. Plant Physiol. 2005. PMID: 16172093 Free PMC article. Review. No abstract available.
-
Effect of temperature on geminivirus-induced RNA silencing in plants.Plant Physiol. 2005 Aug;138(4):1828-41. doi: 10.1104/pp.105.066563. Epub 2005 Jul 22. Plant Physiol. 2005. PMID: 16040661 Free PMC article.
References
-
- Baulcombe, D. (2002) Curr. Biol. 12, R82-R84. - PubMed
-
- Nishikura, K. (2001) Cell 107, 415-418. - PubMed
-
- Waterhouse, P. M., Wang, M.-B. & Lough, T. (2001) Nature 411, 834-842. - PubMed
-
- Tijsterman, M., Ketting, R. F. & Plasterk, R. H. A. (2002) Annu. Rev. Genet. 36, 489-519. - PubMed
-
- Cogoni, C. & Macino, G. (1999) Nature 399, 166-169. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Research Materials

