Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor
- PMID: 15078936
- PMCID: PMC387703
- DOI: 10.1128/jvi.78.9.4552-4560.2004
Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor
Abstract
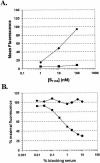
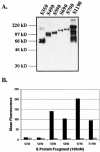
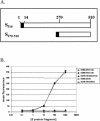
A novel coronavirus, severe acute respiratory syndrome coronavirus (SARS-CoV), has recently been identified as the causative agent of severe acute respiratory syndrome (SARS). SARS-CoV appears similar to other coronaviruses in both virion structure and genome organization. It is known for other coronaviruses that the spike (S) glycoprotein is required for both viral attachment to permissive cells and for fusion of the viral envelope with the host cell membrane. Here we describe the construction and expression of a soluble codon-optimized SARS-CoV S glycoprotein comprising the first 1,190 amino acids of the native S glycoprotein (S(1190)). The codon-optimized and native S glycoproteins exhibit similar molecular weight as determined by Western blot analysis, indicating that synthetic S glycoprotein is modified correctly in a mammalian expression system. S(1190) binds to the surface of Vero E6 cells, a cell permissive to infection, as demonstrated by fluorescence-activated cell sorter analysis, suggesting that S(1190) maintains the biologic activity present in native S glycoprotein. This interaction is blocked with serum obtained from recovering SARS patients, indicating that the binding is specific. In an effort to map the ligand-binding domain of the SARS-CoV S glycoprotein, carboxy- and amino-terminal truncations of the S(1190) glycoprotein were constructed. Amino acids 270 to 510 were the minimal receptor-binding region of the SARS-CoV S glycoprotein as determined by flow cytometry. We speculate that amino acids 1 to 510 of the SARS-CoV S glycoprotein represent a unique domain containing the receptor-binding site (amino acids 270 to 510), analogous to the S1 subunit of other coronavirus S glycoproteins.
Figures
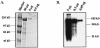



Similar articles
-
Genetic analysis of the SARS-coronavirus spike glycoprotein functional domains involved in cell-surface expression and cell-to-cell fusion.Virology. 2005 Oct 25;341(2):215-30. doi: 10.1016/j.virol.2005.06.046. Epub 2005 Aug 15. Virology. 2005. PMID: 16099010 Free PMC article.
-
Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus.Nature. 2003 Nov 27;426(6965):450-4. doi: 10.1038/nature02145. Nature. 2003. PMID: 14647384 Free PMC article.
-
Antigenicity and receptor-binding ability of recombinant SARS coronavirus spike protein.Biochem Biophys Res Commun. 2004 Jan 23;313(4):938-47. doi: 10.1016/j.bbrc.2003.11.180. Biochem Biophys Res Commun. 2004. PMID: 14706633 Free PMC article.
-
Severe acute respiratory syndrome coronavirus entry as a target of antiviral therapies.Antivir Ther. 2007;12(4 Pt B):639-50. Antivir Ther. 2007. PMID: 17944271 Review.
-
The SARS-CoV S glycoprotein.Cell Mol Life Sci. 2004 Oct;61(19-20):2428-30. doi: 10.1007/s00018-004-4257-y. Cell Mol Life Sci. 2004. PMID: 15526150 Free PMC article. Review.
Cited by
-
Correlation between bioactive lipids and novel coronavirus: constructive role of biolipids in curbing infectivity by enveloped viruses, centralizing on EPA and DHA.Syst Microbiol Biomanuf. 2021;1(2):186-192. doi: 10.1007/s43393-020-00019-3. Epub 2021 Feb 3. Syst Microbiol Biomanuf. 2021. PMID: 38624677 Free PMC article. Review.
-
Comparative genomics of spike, envelope, and nucleocapsid protein of severe acute respiratory syndrome coronavirus 2.Afr Health Sci. 2023 Sep;23(3):384-399. doi: 10.4314/ahs.v23i3.45. Afr Health Sci. 2023. PMID: 38357143 Free PMC article.
-
Chemical and spectroscopic characterization of (Artemisinin/Querctin/ Zinc) novel mixed ligand complex with assessment of its potent high antiviral activity against SARS-CoV-2 and antioxidant capacity against toxicity induced by acrylamide in male rats.PeerJ. 2024 Jan 2;12:e15638. doi: 10.7717/peerj.15638. eCollection 2024. PeerJ. 2024. PMID: 38188145 Free PMC article.
-
Mapping immunological and host receptor binding determinants of SARS-CoV spike protein utilizing the Qubevirus platform.J Biol Chem. 2023 Dec;299(12):105460. doi: 10.1016/j.jbc.2023.105460. Epub 2023 Nov 15. J Biol Chem. 2023. PMID: 37977224 Free PMC article.
-
Characterization of CCoV-HuPn-2018 spike protein-mediated viral entry.J Virol. 2023 Sep 28;97(9):e0060123. doi: 10.1128/jvi.00601-23. Epub 2023 Sep 7. J Virol. 2023. PMID: 37676001 Free PMC article.
References
-
- Bikker, J. A., S. Trumpp-Kallmeyer, and C. Humblet. 1998. G-protein coupled receptors: models, mutagenesis, and drug design. J. Med. Chem. 41:2911-2927. - PubMed
-
- Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

