An overview of gibberellin metabolism enzyme genes and their related mutants in rice
- PMID: 15075394
- PMCID: PMC419838
- DOI: 10.1104/pp.103.033696
An overview of gibberellin metabolism enzyme genes and their related mutants in rice
Erratum in
- Plant Physiol.2004 Jul;135(3):1863
Abstract
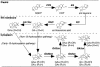
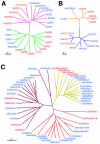
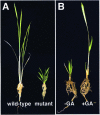
To enhance our understanding of GA metabolism in rice (Oryza sativa), we intensively screened and identified 29 candidate genes encoding the following GA metabolic enzymes using all available rice DNA databases: ent-copalyl diphosphate synthase (CPS), ent-kaurene synthase (KS), ent-kaurene oxidase (KO), ent-kaurenoic acid oxidase (KAO), GA 20-oxidase (GA20ox), GA 3-oxidase (GA3ox), and GA 2-oxidase (GA2ox). In contrast to the Arabidopsis genome, multiple CPS-like, KS-like, and KO-like genes were identified in the rice genome, most of which are contiguously arranged. We also identified 18 GA-deficient rice mutants at six different loci from rice mutant collections. Based on the mutant and expression analyses, we demonstrated that the enzymes catalyzing the early steps in the GA biosynthetic pathway (i.e. CPS, KS, KO, and KAO) are mainly encoded by single genes, while those for later steps (i.e. GA20ox, GA3ox, and GA2ox) are encoded by gene families. The remaining CPS-like, KS-like, and KO-like genes were likely to be involved in the biosynthesis of diterpene phytoalexins rather than GAs because the expression of two CPS-like and three KS-like genes (OsCPS2, OsCPS4, OsKS4, OsKS7, and OsKS8) were increased by UV irradiation, and four of these genes (OsCPS2, OsCPS4, OsKS4, and OsKS7) were also induced by an elicitor treatment.
Figures





Similar articles
-
Genome-wide identification of gibberellins metabolic enzyme genes and expression profiling analysis during seed germination in maize.Gene. 2011 Aug 15;482(1-2):34-42. doi: 10.1016/j.gene.2011.05.008. Epub 2011 May 24. Gene. 2011. PMID: 21640170
-
Transcripts of two ent-copalyl diphosphate synthase genes differentially localize in rice plants according to their distinct biological roles.J Exp Bot. 2015 Jan;66(1):369-76. doi: 10.1093/jxb/eru424. Epub 2014 Oct 21. J Exp Bot. 2015. PMID: 25336684 Free PMC article.
-
Functional characterization of ent-copalyl diphosphate synthase, kaurene synthase and kaurene oxidase in the Salvia miltiorrhiza gibberellin biosynthetic pathway.Sci Rep. 2016 Mar 14;6:23057. doi: 10.1038/srep23057. Sci Rep. 2016. PMID: 26971881 Free PMC article.
-
Evolution of Labdane-Related Diterpene Synthases in Cereals.Plant Cell Physiol. 2020 Dec 23;61(11):1850-1859. doi: 10.1093/pcp/pcaa106. Plant Cell Physiol. 2020. PMID: 32810270 Review.
-
Biosynthesis of gibberellins in Gibberella fujikuroi: biomolecular aspects.Appl Microbiol Biotechnol. 1999 Sep;52(3):298-310. doi: 10.1007/s002530051524. Appl Microbiol Biotechnol. 1999. PMID: 10531641 Review.
Cited by
-
Tomato SlDREB gene restricts leaf expansion and internode elongation by downregulating key genes for gibberellin biosynthesis.J Exp Bot. 2012 Nov;63(18):6407-20. doi: 10.1093/jxb/ers295. Epub 2012 Oct 17. J Exp Bot. 2012. PMID: 23077200 Free PMC article.
-
Phenotypic characterization, genetic mapping and candidate gene analysis of a source conferring reduced plant height in sunflower.Theor Appl Genet. 2013 Jan;126(1):251-63. doi: 10.1007/s00122-012-1978-4. Epub 2012 Sep 13. Theor Appl Genet. 2013. PMID: 22972203
-
Gibberellin Enhances the Anisotropy of Cell Expansion in the Growth Zone of the Maize Leaf.Front Plant Sci. 2020 Aug 4;11:1163. doi: 10.3389/fpls.2020.01163. eCollection 2020. Front Plant Sci. 2020. PMID: 32849718 Free PMC article.
-
Co-inoculations of bacteria and mycorrhizal fungi often drive additive plant growth responses.ISME Commun. 2024 Aug 7;4(1):ycae104. doi: 10.1093/ismeco/ycae104. eCollection 2024 Jan. ISME Commun. 2024. PMID: 39188310 Free PMC article.
-
Functional characterization of wheat ent-kaurene(-like) synthases indicates continuing evolution of labdane-related diterpenoid metabolism in the cereals.Phytochemistry. 2012 Dec;84:47-55. doi: 10.1016/j.phytochem.2012.08.021. Epub 2012 Sep 22. Phytochemistry. 2012. PMID: 23009879 Free PMC article.
References
-
- Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Genet Genomics 267: 730–745 - PubMed
-
- Ashikari M, Sasaki A, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, et al (2002) Loss-of-function of a rice gibberellin biosynthetic gene, GA20 oxidase (GA20ox-2), led to the rice ‘Green Revolution.’ Breed Sci 52: 143–150
-
- Cho E-M, Okada A, Kenmoku H, Otomo K, Toyomasu T, Mitsuhashi W, Sassa T, Yajima A, Yabuta G, Mori K, et al (2004) Molecular cloning and characterization of a cDNA encoding ent-cassa-12,15-diene synthase, a putative diterpenoid phytoalexin biosynthetic enzyme, from suspension-cultured rice cells treated with a chitin elicitor. Plant J 37: 1–8 - PubMed
-
- Evans LT (1993) Crop Evolution, Adaptation and Yield. Cambridge University Press, Cambridge, UK
-
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, Jia P, Zhang Y, et al (2002) Sequence and analysis of rice chromosome 4. Nature 420: 316–320 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

