Molecular clamp mechanism of substrate binding by hydrophobic coiled-coil residues of the archaeal chaperone prefoldin
- PMID: 15070724
- PMCID: PMC384753
- DOI: 10.1073/pnas.0306276101
Molecular clamp mechanism of substrate binding by hydrophobic coiled-coil residues of the archaeal chaperone prefoldin
Abstract
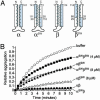
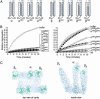
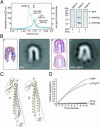
Prefoldin (PFD) is a jellyfish-shaped molecular chaperone that has been proposed to play a general role in de novo protein folding in archaea and is known to assist the biogenesis of actins, tubulins, and potentially other proteins in eukaryotes. Using point mutants, chimeras, and intradomain swap variants, we show that the six coiled-coil tentacles of archaeal PFD act in concert to bind and stabilize nonnative proteins near the opening of the cavity they form. Importantly, the interaction between chaperone and substrate depends on the mostly buried interhelical hydrophobic residues of the coiled coils. We also show by electron microscopy that the tentacles can undergo an en bloc movement to accommodate an unfolded substrate. Our data reveal how archael PFD uses its unique architecture and intrinsic coiled-coil properties to interact with nonnative polypeptides.
Figures

Similar articles
-
Structure and molecular dynamics simulation of archaeal prefoldin: the molecular mechanism for binding and recognition of nonnative substrate proteins.J Mol Biol. 2008 Feb 29;376(4):1130-41. doi: 10.1016/j.jmb.2007.12.010. Epub 2007 Dec 8. J Mol Biol. 2008. PMID: 18201719
-
Divergent substrate-binding mechanisms reveal an evolutionary specialization of eukaryotic prefoldin compared to its archaeal counterpart.Structure. 2007 Jan;15(1):101-10. doi: 10.1016/j.str.2006.11.006. Structure. 2007. PMID: 17223536
-
Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins.Cell. 2000 Nov 10;103(4):621-32. doi: 10.1016/s0092-8674(00)00165-3. Cell. 2000. PMID: 11106732
-
Coiled coils meet the chaperone world.Trends Biochem Sci. 2004 Sep;29(9):455-8. doi: 10.1016/j.tibs.2004.07.004. Trends Biochem Sci. 2004. PMID: 15337117 Review.
-
Structure and function of archaeal prefoldin, a co-chaperone of group II chaperonin.Front Biosci (Landmark Ed). 2010 Jan 1;15(2):708-17. doi: 10.2741/3641. Front Biosci (Landmark Ed). 2010. PMID: 20036841 Review.
Cited by
-
Hold me tight: Role of the heat shock protein family of chaperones in cardiac disease.Circulation. 2010 Oct 26;122(17):1740-51. doi: 10.1161/CIRCULATIONAHA.110.942250. Circulation. 2010. PMID: 20975010 Free PMC article. Review. No abstract available.
-
Prefoldin, a jellyfish-like molecular chaperone: functional cooperation with a group II chaperonin and beyond.Biophys Rev. 2018 Apr;10(2):339-345. doi: 10.1007/s12551-018-0400-0. Epub 2018 Feb 9. Biophys Rev. 2018. PMID: 29427249 Free PMC article. Review.
-
Functional interaction between phosducin-like protein 2 and cytosolic chaperonin is essential for cytoskeletal protein function and cell cycle progression.Mol Biol Cell. 2007 Jun;18(6):2336-45. doi: 10.1091/mbc.e07-01-0069. Epub 2007 Apr 11. Mol Biol Cell. 2007. PMID: 17429077 Free PMC article.
-
A comprehensive analysis of prefoldins and their implication in cancer.iScience. 2021 Oct 15;24(11):103273. doi: 10.1016/j.isci.2021.103273. eCollection 2021 Nov 19. iScience. 2021. PMID: 34761191 Free PMC article. Review.
-
UXT is a novel centrosomal protein essential for cell viability.Mol Biol Cell. 2005 Dec;16(12):5857-65. doi: 10.1091/mbc.e05-08-0705. Epub 2005 Oct 12. Mol Biol Cell. 2005. PMID: 16221885 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

