Physical and functional association of RNA polymerase II and the proteasome
- PMID: 15069196
- PMCID: PMC395896
- DOI: 10.1073/pnas.0305411101
Physical and functional association of RNA polymerase II and the proteasome
Abstract
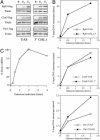
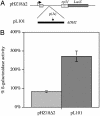
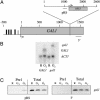
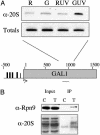
Recent studies from a number of laboratories have revealed a surprising number of connections between RNA polymerase II transcription and the ubiquitin/proteasome pathway. We now find yet another intersection of these pathways by showing that the 26S proteasome associates with regions of the GAL1, GAL10, and HSP82 genes, including the 3' ends, in a transcription-dependent fashion. The appearance of the proteasome on these inducible genes correlates with both the accumulation of transcripts and the buildup of RNA polymerase II complexes in the same region. Furthermore, the 26S proteasome and RNA polymerase II coimmunoprecipitate, and inhibition of 26S proteolytic activity leads to increased read through of a transcription termination site. We suggest that the proteasome is generally recruited to the DNA at sites of stalled RNA polymerase and may act to resolve these complexes.
Figures


Similar articles
-
Demonstration that a human 26S proteolytic complex consists of a proteasome and multiple associated protein components and hydrolyzes ATP and ubiquitin-ligated proteins by closely linked mechanisms.Eur J Biochem. 1992 Jun 1;206(2):567-78. doi: 10.1111/j.1432-1033.1992.tb16961.x. Eur J Biochem. 1992. PMID: 1317798
-
Regulation of proteolysis.Curr Opin Clin Nutr Metab Care. 2001 Jan;4(1):45-9. doi: 10.1097/00075197-200101000-00009. Curr Opin Clin Nutr Metab Care. 2001. PMID: 11122559 Review.
-
The 26S proteasome: a molecular machine designed for controlled proteolysis.Annu Rev Biochem. 1999;68:1015-68. doi: 10.1146/annurev.biochem.68.1.1015. Annu Rev Biochem. 1999. PMID: 10872471 Review.
-
Emerging roles of ubiquitin in transcription regulation.Science. 2002 May 17;296(5571):1254-8. doi: 10.1126/science.1067466. Science. 2002. PMID: 12016299 Review.
-
The proteasome.Curr Biol. 1998 Dec 17-31;8(25):R902. doi: 10.1016/s0960-9822(98)00004-9. Curr Biol. 1998. PMID: 9889109 Review. No abstract available.
Cited by
-
Stuxnet Facilitates the Degradation of Polycomb Protein during Development.Dev Cell. 2016 Jun 20;37(6):507-19. doi: 10.1016/j.devcel.2016.05.013. Dev Cell. 2016. PMID: 27326929 Free PMC article.
-
A SUMO-dependent pathway controls elongating RNA Polymerase II upon UV-induced damage.Sci Rep. 2019 Nov 29;9(1):17914. doi: 10.1038/s41598-019-54027-y. Sci Rep. 2019. PMID: 31784551 Free PMC article.
-
Ubp10/Dot4p regulates the persistence of ubiquitinated histone H2B: distinct roles in telomeric silencing and general chromatin.Mol Cell Biol. 2005 Jul;25(14):6123-39. doi: 10.1128/MCB.25.14.6123-6139.2005. Mol Cell Biol. 2005. PMID: 15988024 Free PMC article.
-
Toward accurate reconstruction of functional protein networks.Mol Syst Biol. 2009;5:248. doi: 10.1038/msb.2009.3. Epub 2009 Mar 17. Mol Syst Biol. 2009. PMID: 19293828 Free PMC article.
-
Cks1 activates transcription by binding to the ubiquitylated proteasome.Mol Cell Biol. 2010 Aug;30(15):3894-901. doi: 10.1128/MCB.00655-09. Epub 2010 Jun 1. Mol Cell Biol. 2010. PMID: 20516216 Free PMC article.
References
-
- Baumeister, W., Walz, J., Zuhl, F. & Seemuller, E. (1998) Cell 92, 367–380. - PubMed
-
- Gonzalez, F., Delahodde, A., Kodadek, T. & Johnston, S. A. (2002) Science 296, 548–550. - PubMed
-
- Ferdous, A., Gonzalez, F., Sun, L., Kodadek, T. & Johnston, S. A. (2001) Mol. Cell 7, 981–991. - PubMed
-
- Ferdous, A., Kodadek, T. & Johnston, S. A. (2002) Biochemistry 41, 12798–12805. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

