Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors
- PMID: 15043961
- PMCID: PMC7140173
- DOI: 10.1016/S0140-6736(04)15788-7
Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors
Abstract
Background: Studies on the fusion-inhibitory peptides derived from the heptad repeat 1 and 2 (HR1 and HR2) regions of the HIV-1 envelope glycoprotein gp41 provided crucial information on the viral fusogenic mechanism. We used a similar approach to study the fusogenic mechanism of severe-acute-respiratory-syndrome-associated coronavirus (SARS-CoV).
Methods: We tested the inhibitory activity against infection of two sets of peptides corresponding to sequences of SARS-CoV spike protein HR1 and HR2 regions and investigated the interactions between the HR1 and HR2 peptides by surface plasmon resonance, sedimentation equilibration analysis, circular dichroism, native polyacrylamide-gel electrophoresis, size exclusion high-performance liquid chromatography, and computer-aided homology modelling and molecule docking analysis.
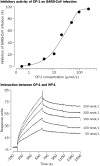
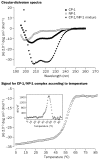
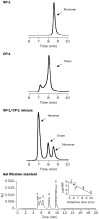
Findings: One peptide, CP-1, derived from the HR2 region, inhibited SARS-CoV infection in the micromolar range. CP-1 bound with high affinity to a peptide from the HR1 region, NP-1. CP-1 alone had low alpha-helicity and self-associated to form a trimer in phosphate buffer (pH 7.2). CP-1 and NP-1 mixed in equimolar concentrations formed a six-helix bundle, similar to the fusogenic core structure of HIV-1 gp41.
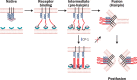
Interpretation: After binding to the target cell, the transmembrane spike protein might change conformation by association between the HR1 and HR2 regions to form an oligomeric structure, leading to fusion between the viral and target-cell membranes. At the prefusion intermediate state, CP-1 could bind to the HR1 region and interfere with the conformational changes, resulting in inhibition of SARS-CoV fusion with the target cells. CP-1 might be modifiable to increase its anti-SARS-CoV activity and could be further developed as an antiviral agent for treatment or prophylaxis of SARS-CoV infection.
Figures



Similar articles
-
Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides.Proc Natl Acad Sci U S A. 2004 Jun 1;101(22):8455-60. doi: 10.1073/pnas.0400576101. Epub 2004 May 18. Proc Natl Acad Sci U S A. 2004. PMID: 15150417 Free PMC article.
-
Solution structure of the severe acute respiratory syndrome-coronavirus heptad repeat 2 domain in the prefusion state.J Biol Chem. 2006 Apr 28;281(17):11965-71. doi: 10.1074/jbc.M601174200. Epub 2006 Feb 28. J Biol Chem. 2006. PMID: 16507566 Free PMC article.
-
Identification of a minimal peptide derived from heptad repeat (HR) 2 of spike protein of SARS-CoV and combination of HR1-derived peptides as fusion inhibitors.Antiviral Res. 2009 Jan;81(1):82-7. doi: 10.1016/j.antiviral.2008.10.001. Epub 2008 Nov 5. Antiviral Res. 2009. PMID: 18983873 Free PMC article.
-
Multimerization of the heptad repeat regions of the SARS-CoV 2 spike protein.Biochim Biophys Acta Biomembr. 2024 Feb;1866(2):184259. doi: 10.1016/j.bbamem.2023.184259. Epub 2023 Dec 5. Biochim Biophys Acta Biomembr. 2024. PMID: 38061554 Review.
-
Severe acute respiratory syndrome coronavirus entry into host cells: Opportunities for therapeutic intervention.Med Res Rev. 2006 Jul;26(4):414-33. doi: 10.1002/med.20055. Med Res Rev. 2006. PMID: 16521129 Free PMC article. Review.
Cited by
-
Monitoring correlates of SARS-CoV-2 infection in cell culture using a two-photon-active calcium-sensitive dye.Cell Mol Biol Lett. 2024 Jul 19;29(1):105. doi: 10.1186/s11658-024-00619-0. Cell Mol Biol Lett. 2024. PMID: 39030477 Free PMC article.
-
The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.Curr Res Struct Biol. 2024 Apr 29;7:100148. doi: 10.1016/j.crstbi.2024.100148. eCollection 2024. Curr Res Struct Biol. 2024. PMID: 38742159 Free PMC article.
-
Beyond acute infection: molecular mechanisms underpinning cardiovascular complications in long COVID.Front Cardiovasc Med. 2024 Feb 14;11:1268571. doi: 10.3389/fcvm.2024.1268571. eCollection 2024. Front Cardiovasc Med. 2024. PMID: 38495940 Free PMC article. Review.
-
An enhanced broad-spectrum peptide inhibits Omicron variants in vivo.Cell Rep Med. 2024 Feb 20;5(2):101418. doi: 10.1016/j.xcrm.2024.101418. Epub 2024 Feb 9. Cell Rep Med. 2024. PMID: 38340726 Free PMC article.
-
Dramatic Differences between the Structural Susceptibility of the S1 Pre- and S2 Postfusion States of the SARS-CoV-2 Spike Protein to External Electric Fields Revealed by Molecular Dynamics Simulations.Viruses. 2023 Dec 11;15(12):2405. doi: 10.3390/v15122405. Viruses. 2023. PMID: 38140646 Free PMC article.
References
-
- WHO Summary of probable SARS cases with onset of illness from Nov 1, 2002, to July 31, 2003. www.who.int (accessed Dec 19, 2003)
-
- Ksiazek TG, Erdman D, Goldsmith CS. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. - PubMed
-
- Drosten C, Gunther S, Preiser W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. NEnglJ Med. 2003;348:1967–1976. - PubMed
-
- Marra MA, Jones SJM, Astell CR. The genome sequence of the SARS-associated coronavirus. Science. 2003;300:1399–1404. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

