Different gene expression patterns in invasive lobular and ductal carcinomas of the breast
- PMID: 15034139
- PMCID: PMC420079
- DOI: 10.1091/mbc.e03-11-0786
Different gene expression patterns in invasive lobular and ductal carcinomas of the breast
Abstract
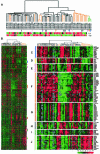
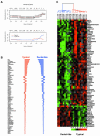
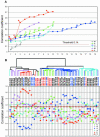
Invasive ductal carcinoma (IDC) and invasive lobular carcinoma (ILC) are the two major histological types of breast cancer worldwide. Whereas IDC incidence has remained stable, ILC is the most rapidly increasing breast cancer phenotype in the United States and Western Europe. It is not clear whether IDC and ILC represent molecularly distinct entities and what genes might be involved in the development of these two phenotypes. We conducted comprehensive gene expression profiling studies to address these questions. Total RNA from 21 ILCs, 38 IDCs, two lymph node metastases, and three normal tissues were amplified and hybridized to approximately 42,000 clone cDNA microarrays. Data were analyzed using hierarchical clustering algorithms and statistical analyses that identify differentially expressed genes (significance analysis of microarrays) and minimal subsets of genes (prediction analysis for microarrays) that succinctly distinguish ILCs and IDCs. Eleven of 21 (52%) of the ILCs ("typical" ILCs) clustered together and displayed different gene expression profiles from IDCs, whereas the other ILCs ("ductal-like" ILCs) were distributed between different IDC subtypes. Many of the differentially expressed genes between ILCs and IDCs code for proteins involved in cell adhesion/motility, lipid/fatty acid transport and metabolism, immune/defense response, and electron transport. Many genes that distinguish typical and ductal-like ILCs are involved in regulation of cell growth and immune response. Our data strongly suggest that over half the ILCs differ from IDCs not only in histological and clinical features but also in global transcription programs. The remaining ILCs closely resemble IDCs in their transcription patterns. Further studies are needed to explore the differences between ILC molecular subtypes and to determine whether they require different therapeutic strategies.
Figures


Similar articles
-
The molecular underpinning of lobular histological growth pattern: a genome-wide transcriptomic analysis of invasive lobular carcinomas and grade- and molecular subtype-matched invasive ductal carcinomas of no special type.J Pathol. 2010 Jan;220(1):45-57. doi: 10.1002/path.2629. J Pathol. 2010. PMID: 19877120
-
Lobular and ductal carcinomas of the breast have distinct genomic and expression profiles.Oncogene. 2008 Sep 11;27(40):5359-72. doi: 10.1038/onc.2008.158. Epub 2008 May 19. Oncogene. 2008. PMID: 18490921 Free PMC article.
-
Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis.BMC Cancer. 2007 Mar 27;7:55. doi: 10.1186/1471-2407-7-55. BMC Cancer. 2007. PMID: 17389037 Free PMC article.
-
Comprehensive Review of Molecular Mechanisms and Clinical Features of Invasive Lobular Cancer.Oncologist. 2021 Jun;26(6):e943-e953. doi: 10.1002/onco.13734. Epub 2021 Mar 16. Oncologist. 2021. PMID: 33641217 Free PMC article. Review.
-
Differentiation of tumours of ductal and lobular origin: II. Genomics of invasive ductal and lobular breast carcinomas.Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2005 Jun;149(1):63-8. doi: 10.5507/bp.2005.006. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2005. PMID: 16170390 Review.
Cited by
-
Transcriptomic analysis identifies enrichment of cAMP/PKA/CREB signaling in invasive lobular breast cancer.Breast Cancer Res. 2024 Oct 30;26(1):149. doi: 10.1186/s13058-024-01900-y. Breast Cancer Res. 2024. PMID: 39478577 Free PMC article.
-
Comprehensive analysis of expression and prognostic value of the claudin family in human breast cancer.Aging (Albany NY). 2021 Mar 10;13(6):8777-8796. doi: 10.18632/aging.202687. Epub 2021 Mar 10. Aging (Albany NY). 2021. PMID: 33714203 Free PMC article.
-
Using Frequent Co-expression Network to Identify Gene Clusters for Breast Cancer Prognosis.Proc Int Joint Conf Bioinforma Syst Biol Intell Comput. 2009 Aug 3:428-434. doi: 10.1109/IJCBS.2009.29. Proc Int Joint Conf Bioinforma Syst Biol Intell Comput. 2009. PMID: 23615925 Free PMC article.
-
Translational pathology of neoplasia.Cancer Biomark. 2010;9(1-6):7-20. doi: 10.3233/CBM-2011-0159. Cancer Biomark. 2010. PMID: 22112467 Free PMC article.
-
Polymorphisms in the gene regions of the adaptor complex LAMTOR2/LAMTOR3 and their association with breast cancer risk.PLoS One. 2013;8(1):e53768. doi: 10.1371/journal.pone.0053768. Epub 2013 Jan 16. PLoS One. 2013. PMID: 23341997 Free PMC article.
References
-
- Agarwal, V.R., Bischoff, E.D., Hermann, T., and Lamph, W.W. (2000). Induction of adipocyte specific gene expression is correlated with mammary tumor regression by the retinoid X receptor-ligand LGD1069 (targretin). Cancer Res. 60, 6033-6038. - PubMed
-
- Ahr, A., Holtrich, U., Solbach, C., Scharl, A., Strebhardt, K., Karn, T., and Kaufmann, M. (2001). Molecular classification of breast cancer patients by gene expression profiling. J. Pathol. 195, 312-320. - PubMed
-
- Ahr, A., Karn, T., Solbach, C., Seiter, T., Strebhardt, K., Holtrich, U., and Kaufmann, M. (2002). Identification of high risk breast-cancer patients by gene expression profiling. Lancet 359, 131-132. - PubMed
-
- Bartsch, H., Nair, J., and Owen, R.W. (1999). Dietary polyunsaturated fatty acids and cancers of the breast and colorectum: emerging evidence for their role as risk modifiers. Carcinogenesis 20, 2209-2218. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

