Preferential duplication of conserved proteins in eukaryotic genomes
- PMID: 15024414
- PMCID: PMC368158
- DOI: 10.1371/journal.pbio.0020055
Preferential duplication of conserved proteins in eukaryotic genomes
Abstract
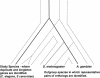
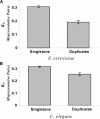
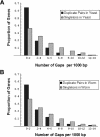
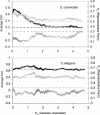
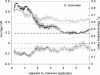
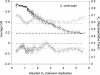
A central goal in genome biology is to understand the origin and maintenance of genic diversity. Over evolutionary time, each gene's contribution to the genic content of an organism depends not only on its probability of long-term survival, but also on its propensity to generate duplicates that are themselves capable of long-term survival. In this study we investigate which types of genes are likely to generate functional and persistent duplicates. We demonstrate that genes that have generated duplicates in the C. elegans and S. cerevisiae genomes were 25%-50% more constrained prior to duplication than the genes that failed to leave duplicates. We further show that conserved genes have been consistently prolific in generating duplicates for hundreds of millions of years in these two species. These findings reveal one way in which gene duplication shapes the content of eukaryotic genomes. Our finding that the set of duplicate genes is biased has important implications for genome-scale studies.
Conflict of interest statement
The authors have declared that no conflicts of interest exist.
Figures
Similar articles
-
The evolutionary fate and consequences of duplicate genes.Science. 2000 Nov 10;290(5494):1151-5. doi: 10.1126/science.290.5494.1151. Science. 2000. PMID: 11073452
-
Variation in gene duplicates with low synonymous divergence in Saccharomyces cerevisiae relative to Caenorhabditis elegans.Genome Biol. 2009;10(7):R75. doi: 10.1186/gb-2009-10-7-r75. Epub 2009 Jul 13. Genome Biol. 2009. PMID: 19594930 Free PMC article.
-
Comparison of the oxidative phosphorylation (OXPHOS) nuclear genes in the genomes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae.Genome Biol. 2005;6(2):R11. doi: 10.1186/gb-2005-6-2-r11. Epub 2005 Jan 31. Genome Biol. 2005. PMID: 15693940 Free PMC article.
-
Detection of gene duplications and block duplications in eukaryotic genomes.J Struct Funct Genomics. 2003;3(1-4):27-34. J Struct Funct Genomics. 2003. PMID: 12836682 Review.
-
Birth and death of duplicated genes in completely sequenced eukaryotes.Trends Genet. 2001 May;17(5):237-9. doi: 10.1016/s0168-9525(01)02243-0. Trends Genet. 2001. PMID: 11335019 Review.
Cited by
-
Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants.Plant Physiol. 2013 Apr;161(4):1844-61. doi: 10.1104/pp.112.200147. Epub 2013 Feb 8. Plant Physiol. 2013. PMID: 23396833 Free PMC article.
-
How much does the amphioxus genome represent the ancestor of chordates?Brief Funct Genomics. 2012 Mar;11(2):89-95. doi: 10.1093/bfgp/els003. Epub 2012 Feb 28. Brief Funct Genomics. 2012. PMID: 22373648 Free PMC article. Review.
-
Gene duplication and environmental adaptation within yeast populations.Genome Biol Evol. 2010;2:591-601. doi: 10.1093/gbe/evq043. Epub 2010 Jul 21. Genome Biol Evol. 2010. PMID: 20660110 Free PMC article.
-
Asymmetric and non-uniform evolution of recently duplicated human genes.Biol Direct. 2010 Sep 8;5:54. doi: 10.1186/1745-6150-5-54. Biol Direct. 2010. PMID: 20825637 Free PMC article.
-
Dynamic evolution of megasatellites in yeasts.Nucleic Acids Res. 2010 Aug;38(14):4731-9. doi: 10.1093/nar/gkq207. Epub 2010 Mar 31. Nucleic Acids Res. 2010. PMID: 20360043 Free PMC article.
References
-
- Akashi H. Gene expression and molecular evolution. Curr Opin Genet Dev. 2001;11:660–666. - PubMed
-
- Bromham L, Penny D. The modern molecular clock. Nat Rev Genet. 2003;4:216–224. - PubMed
-
- Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–942. - PubMed
-
- Carbone A, Zinovyev A, Kepes F. Codon adaptaion index as a measure of dominating codon bias. Bioinformatics. 2003;19:2005–2015. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

